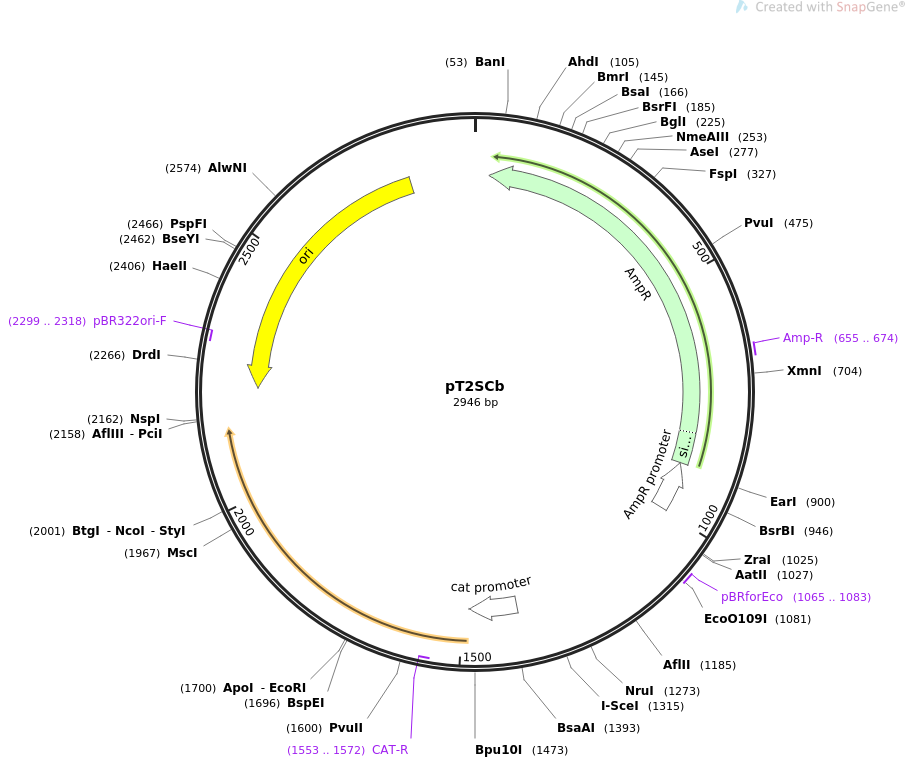
pT2SCb
(Plasmid
#59385)
-
PurposeTemplate plasmid for amplification of a selection cassette that includes two transcription terminator elements, an I-SceI site and a modified chloramphenicol resistance gene.
-
Depositing Lab
-
Publication
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 59385 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
This material is available to academics and nonprofits only.
Backbone
-
Vector backbonepHA1887, an 1887 bp fragment extending from bp 690 to bp 2576 of pUC19 (Acc. No. M77789)
-
Backbone manufacturerCopley Lab
- Backbone size w/o insert (bp) 1887
- Total vector size (bp) 2946
-
Vector typetemplate
Growth in Bacteria
-
Bacterial Resistance(s)Chloramphenicol and Ampicillin, 25 & 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)Mach1
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameTT-ISceI-cat2
-
SpeciesSynthetic
-
Insert Size (bp)1059
-
MutationThe 3’-end of the chloramphenicol resistance gene was modified to 5’-AGGAGGTGCATAA to create a sequence resembling the consensus bacterial ribosomal binding site (5’-AGGAGGTAAATAA).
-
GenBank IDKP897157.1
Cloning Information
- Cloning method Gibson Cloning
- 5′ sequencing primer tatcagggttattgtctcatg
- 3′ sequencing primer acttgagcgtcgatttttgtg (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
iGEM parts BBa_B1002 and BBa_B1006 were used to build the double terminator element. The chloramphenicol resistance gene was taken from pACYC187. The 3'-end was modified from 5'-GGGCGGGGCGTAA to 5'-AGGAGGTGCATAA in order to provide a site resembling the consensus bacterial ribosomal binding site (5'-AGGAGGTAAATAA). We developed GetX (https://sourceforge.net/projects/getx/), a stand-alone python script that allows the user to design mutation cassettes for scarless genome editing in bacteria using our previously described two-step recombination method. Please see the document linked under the Resource Information heading above for additional information on installing and using the script.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pT2SCb was a gift from Shelley Copley (Addgene plasmid # 59385 ; http://n2t.net/addgene:59385 ; RRID:Addgene_59385) -
For your References section:
A versatile and highly efficient method for scarless genome editing in Escherichia coli and Salmonella enterica. Kim J, Webb AM, Kershner JP, Blaskowski S, Copley SD. BMC Biotechnol. 2014 Sep 25;14(1):84. 10.1186/1472-6750-14-84 PubMed 25255806