We narrowed to 39,979 results for: KAN
-
Plasmid#195947PurposeTp901 target with pUBQ10 switching from mTurq to mScarletDepositorInserttRBCS-mScarlet-attP_Tp901-pUBQ10-attB_Tp901-mTurq-tUBQ10
ExpressionPlantAvailable SinceMarch 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
T1B/pUBQ10(mTurq->mScarlet)
Plasmid#195946PurposeBxb1 target with pUBQ10 switching from mTurq to mScarletDepositorInserttRBCS-mScarlet-attP_Bxb1-pUBQ10-attB_Bxb1-mTurq-tUBQ10
ExpressionPlantAvailable SinceMarch 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
pMuSIC pool v1.0
Pooled Library#227193PurposeTwo-way barcodes from 18 fluorescent proteins for mammalian expression. Can be used for clone tracing, or for pairing with perturbations for single cell resolution in spectral detection measurements.DepositorExpressionMammalianAvailable SinceDec. 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library A
Pooled Library#157971PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
Ma-PylRS-lib1
Pooled Library#224585PurposePyrrolysine tRNA synthetase library to generate new genetic code expansion (GCE) systems for site-specific incorporation of non-canonical amino acids (ncAAs) into proteins.DepositorExpressionBacterialSpeciesMethanomethylophilus alvusAvailable SinceJan. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library 1
Pooled Library#168776PurposePooled library expressing barcoded SARS-CoV-2 RNA Binding Domain mutationsDepositorExpressionYeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library 2
Pooled Library#168777PurposePooled library expressing barcoded SARS-CoV-2 RNA Binding Domain mutationsDepositorExpressionYeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
pTet-IRES-EGFP
Plasmid#64238PurposeLentiviral, doxycycline-inducible system to express transgene of interest and EGFP.DepositorHas ServiceCloning Grade DNATypeEmpty backboneUseCre/Lox and LentiviralExpressionMammalianPromoterpTetAvailable SinceJune 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
pMJA284= pSin-Pur-MCS-Puro
Plasmid#116877PurposeLentiviral transfer vector containing a multible cloning siteDepositorTypeEmpty backboneUseLentiviralExpressionMammalianAvailable SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
pL0R_lacZ
Plasmid#202095PurposeReceiver cloning vector for making level 0 parts. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pBbB8k-csg-amylase
Plasmid#166859PurposeArabinose-inducible csgBACEFG operon for curli fiber synthesis in which curli subunit CsgA has been fused to α-amylase via a flexible linker. Derived from https://www.addgene.org/35363.DepositorInsertcsgBACEFG, with α-amylase fused to CsgA via flexible linker
UseSynthetic BiologyExpressionBacterialAvailable SinceApril 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
HR700_TP53Exon5mut
Pooled Library#229137PurposeDonor vector library to introduce TP53 mutations into the endogenous gene locus at exon 5 of human cells via CRISPR-HDR.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and Cre/LoxAvailable SinceFeb. 24, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
HR700_TP53Exon6mut
Pooled Library#229138PurposeDonor vector library to introduce TP53 mutations into the endogenous gene locus at exon 6 of human cells via CRISPR-HDR.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and Cre/LoxAvailable SinceFeb. 24, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
HR700_TP53Exon7mut
Pooled Library#229139PurposeDonor vector library to introduce TP53 mutations into the endogenous gene locus at exon 7 of human cells via CRISPR-HDR.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and Cre/LoxAvailable SinceFeb. 24, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
HR700_TP53Exon8mut
Pooled Library#229140PurposeDonor vector library to introduce TP53 mutations into the endogenous gene locus at exon 8 of human cells via CRISPR-HDR.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and Cre/LoxAvailable SinceFeb. 24, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
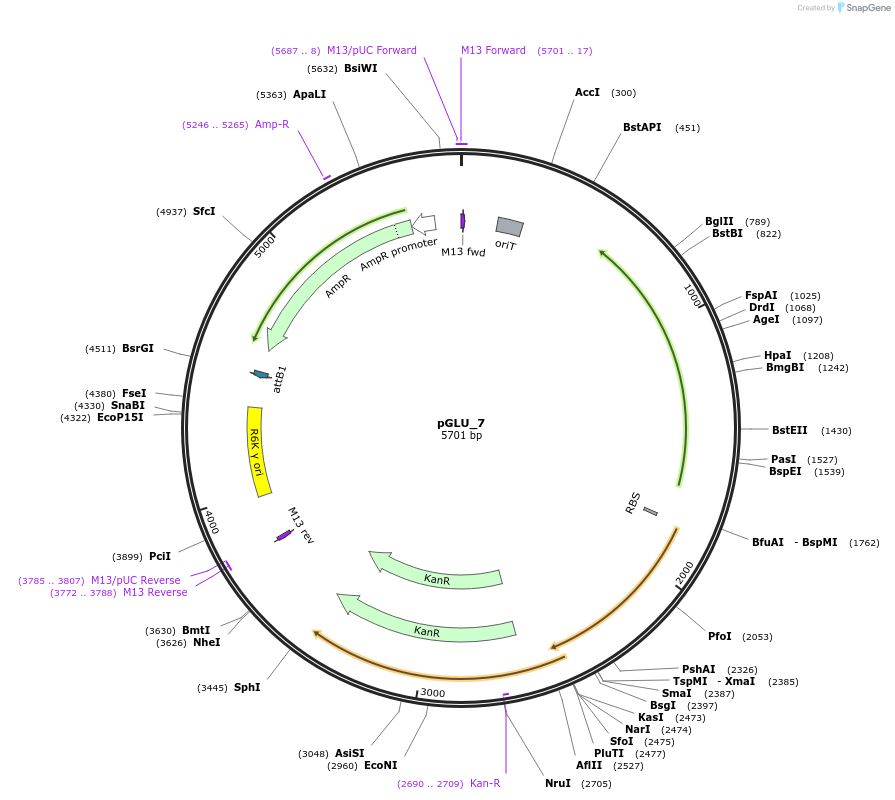
pGLU_7
Plasmid#225815PurposeTn libraries Himar variant KAN backboneDepositorTypeEmpty backboneUseSynthetic BiologyMutationN/AAvailable SinceOct. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
pGLU_128
Plasmid#225821PurposeTn libraries Tn5 variant KAN backboneDepositorTypeEmpty backboneUseSynthetic BiologyMutationN/AAvailable SinceOct. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 2 (positions 437-527)
Pooled Library#174296PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 1 (positions 333-436)
Pooled Library#174295PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 2 (positions 437-527)
Pooled Library#174294PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 1 (positions 333-436)
Pooled Library#174293PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
pBbB8k-csg-NbGFP
Plasmid#166858PurposeArabinose-inducible csgBACEFG operon for curli fiber synthesis in which curli subunit CsgA has been fused to a GFP-specific nanobody via a flexible linker. Derived from https://www.addgene.org/35363/.DepositorInsertcsgBACEFG, with GFP-specific nanobody fused to CsgA via flexible linker
UseSynthetic BiologyExpressionBacterialAvailable SinceMarch 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
M13-kmR
Plasmid#206861PurposeM13KE from NEB modified with kanamycin resistance gene and NotI restriction site on g3p ( M13KE-NotI-Kan)DepositorInsertKanamycin resistance gene
ExpressionBacterialPromoterlacAvailable SinceNov. 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
pCA_o-4
Plasmid#202094PurposeReceiver cloning vector for odd-4 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pCA_o-3
Plasmid#202093PurposeReceiver cloning vector for odd-3 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pCA_o-2
Plasmid#202092PurposeReceiver cloning vector for odd-2 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pCA_e-1
Plasmid#202096PurposeReceiver cloning vector for even-1 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pCA_o-1
Plasmid#202091PurposeReceiver cloning vector for odd-1 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pCA_e-2
Plasmid#202097PurposeReceiver cloning vector for even-2 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pL0_DE_mTurquoise2
Plasmid#202155PurposeLevel 0 plasmid containing a mTurquoise2 fluorescent protein with DE overhangs used to build a level 1 construct.DepositorInsertmTurquoise2 gene
UseSynthetic BiologyMutationNoneAvailable SinceAug. 25, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pCA_e-4
Plasmid#202099PurposeReceiver cloning vector for even-4 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pCA_e-3
Plasmid#202098PurposeReceiver cloning vector for even-3 level constructs. Contains lacZ for blue/white screening.DepositorTypeEmpty backboneUseSynthetic BiologyMutationNoneAvailable SinceAug. 23, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
makeTCR_HDR_Hs.TRAC_Backbone
Plasmid#233944Purposebackbone for makeTCR assemblyDepositorTypeEmpty backboneExpressionMammalianPromoterknock-in to endogenous TRACAvailable SinceApril 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
pT7CFE1-Nhis-GST-CHA-YTH domain (YTHDC1, 317-532)
Plasmid#102277PurposeFor expression of His-GST-tagged human YTHDC1 YTH domain using mammalian cell-free protein expression system (1-step human high-yield mini IVT kit, cat. #88890, Thermo Fisher Scientific)DepositorInsertYTH domain of human YTHDC1 (317-532) (YTHDC1 Human)
UseFor mammalian cell-free protein expression system…TagsGST, HA, and HISPromoterT7Available SinceJuly 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
ML3
Bacterial Strain#61907PurposeC43(DE3)-based E. coli amino acid auxotrophic host strains used for selective isotope labeling. hisG gene deleted.DepositorBacterial ResistanceKanamycinAvailable SinceJan. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
ML8
Bacterial Strain#61909PurposeC43(DE3)-based E. coli amino acid auxotrophic host strains used for selective isotope labeling. argH gene deleted.DepositorBacterial ResistanceKanamycinAvailable SinceJan. 26, 2015AvailabilityAcademic Institutions and Nonprofits only -
pL0_AC-CEN_ARS_HIS
Plasmid#202129PurposeLevel 0 plasmid containing a centromere sequence that is recognized in diatoms with AC overhangs used to build level 1 constructs.DepositorInsertCEN_ARS_HIS
UseSynthetic BiologyMutationARS sequence mutated to remove sapI site for dome…Available SinceAug. 25, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pL0_EF-PtBle
Plasmid#202180PurposeLevel 0 plasmid containing a phleomycin antibiotic cassette (fcpF promoter) with EF overhangs used to build a level 1 construct.DepositorInsertShBle expression cassette
UseSynthetic BiologyMutationNoneAvailable SinceAug. 29, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pL0_EF-H4
Plasmid#202178PurposeLevel 0 plasmid containing a terminator sequence from gene 34971 from Phaeodactylum tricornutum with EF overhangs used to build a level 1 construct.DepositorInsertH4 terminator
UseSynthetic BiologyMutationNoneAvailable SinceAug. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
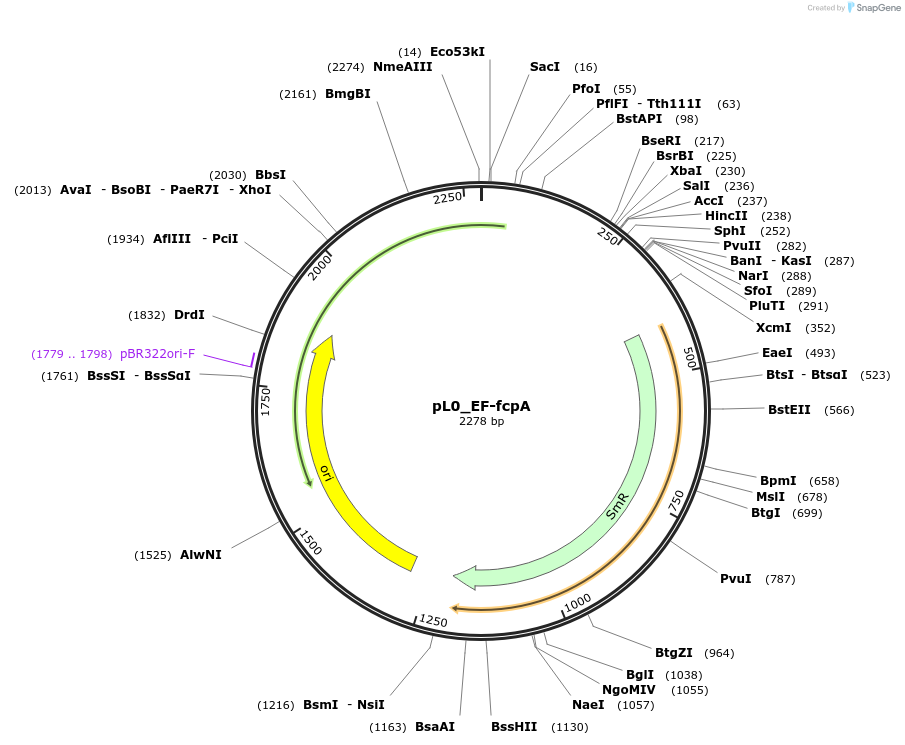
pL0_EF-fcpA
Plasmid#202164PurposeLevel 0 plasmid containing a terminator sequence from gene 18049 from Phaeodactylum tricornutum with EF overhangs used to build a level 1 construct.DepositorInsertfcpA terminator
UseSynthetic BiologyMutationNoneAvailable SinceAug. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pL0_AC-H4
Plasmid#202122PurposeLevel 0 plasmid containing the promoter from gene 34971 from Phaeodactylum tricornutum with AC overhangs used to build a level 1 construct.DepositorInsertH4 promoter
UseSynthetic BiologyMutationNoneAvailable SinceAug. 24, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pL0_AC-P49
Plasmid#202120PurposeLevel 0 plasmid containing the promoter from gene 49202 from Phaeodactylum tricornutum with AC overhangs used to build a level 1 construct.DepositorInsertP49 promoter
UseSynthetic BiologyMutationNoneAvailable SinceAug. 24, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pL0_EF-Fld
Plasmid#202181PurposeLevel 0 plasmid containing a terminator sequence from gene 23658 from Phaeodactylum tricornutum with EF overhangs used to build a level 1 construct.DepositorInsertFld terminator
UseSynthetic BiologyMutationNoneAvailable SinceFeb. 2, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
pL0_EF-P49
Plasmid#202176PurposeLevel 0 plasmid containing a terminator sequence from gene 49202 from Phaeodactylum tricornutum with EF overhangs used to build a level 1 construct.DepositorInsertP49 terminator
UseSynthetic BiologyMutationNoneAvailable SinceOct. 19, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pL0_DE-Per_sig_pep
Plasmid#202152PurposeLevel 0 plasmid containing a peroxisome signal peptide sequence with DE overhangs used to build level 1 constructs.DepositorInsertPeroxisome signal peptide
UseSynthetic BiologyMutationNoneAvailable SinceOct. 19, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pL0_EF-act1_full
Plasmid#202172PurposeLevel 0 plasmid containing a terminator sequence from gene 29812 from Phaeodactylum tricornutum with EF overhangs used to build a level 1 construct.DepositorInsertact1 full terminator
UseSynthetic BiologyMutationNoneAvailable SinceSept. 15, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pL0_EF-glna
Plasmid#202183PurposeLevel 0 plasmid containing a terminator sequence from gene 22357 from Phaeodactylum tricornutum with EF overhangs used to build a level 1 construct.DepositorInsertglna terminator
UseSynthetic BiologyMutationNoneAvailable SinceAug. 29, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
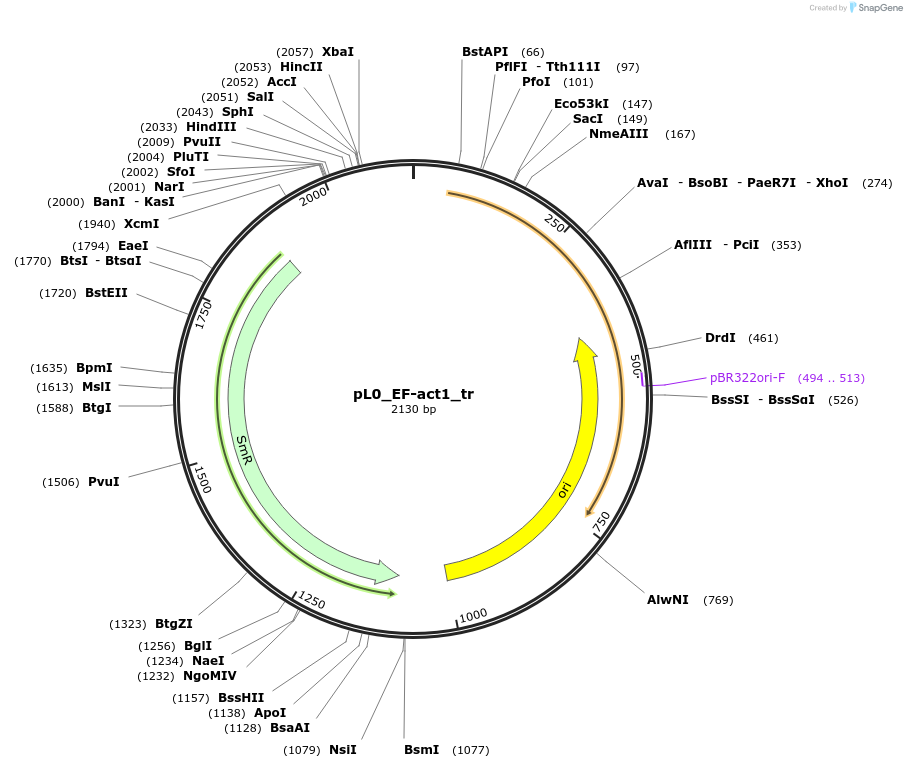
pL0_EF-act1_tr
Plasmid#202171PurposeLevel 0 plasmid containing a terminator sequence from gene 29812 from Phaeodactylum tricornutum with EF overhangs used to build a level 1 construct.DepositorInsertact1 truncated terminator
UseSynthetic BiologyMutationNoneAvailable SinceAug. 29, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pL0_AC-GPI1
Plasmid#202124PurposeLevel 0 plasmid containing the promoter from gene 23924 from Phaeodactylum tricornutum with AC overhangs used to build a level 1 construct.DepositorInsertGPI1 promoter
UseSynthetic BiologyMutationNoneAvailable SinceAug. 29, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
pL0_AC-fcpF
Plasmid#202119PurposeLevel 0 plasmid containing the promoter from gene 51230 from Phaeodactylum tricornutum with AC overhangs used to build a level 1 construct.DepositorInsertfcpF promoter
UseSynthetic BiologyMutationNoneAvailable SinceAug. 29, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits