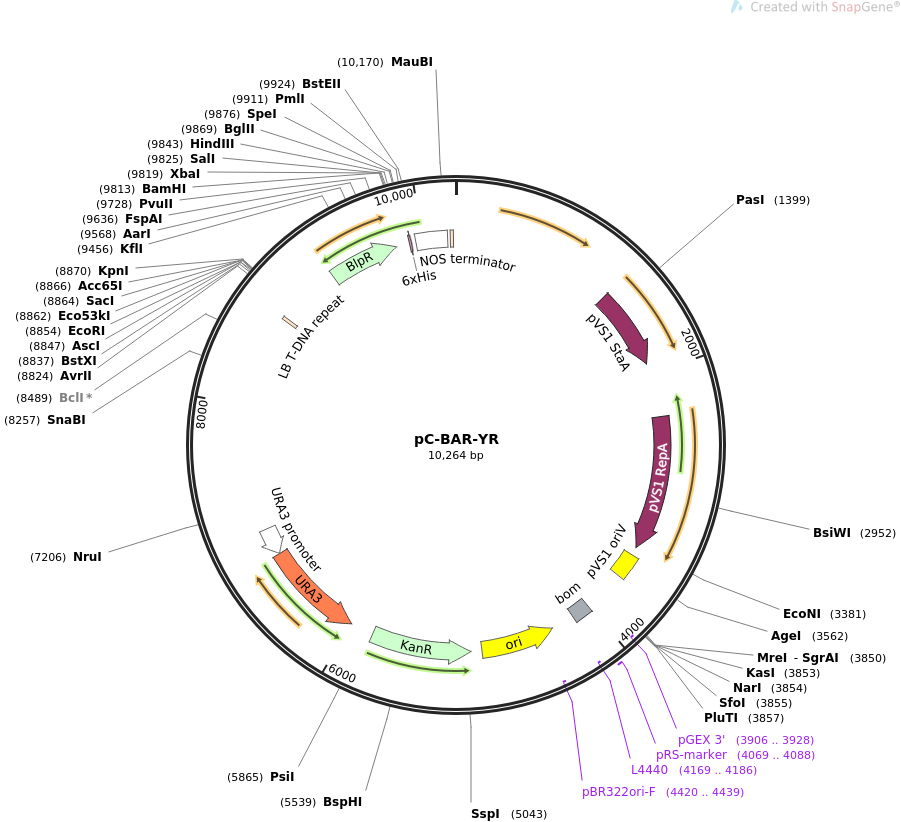
pC-BAR-YR
(Plasmid
#61766)
-
PurposeYeast recombinational cloning compatible Agrobacterium tumefaciens ternary vector containing bar gene (for resistance against glufosinate ammonium) on transfer DNA (TDNA).
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 61766 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
This material is available to academics and nonprofits only.
Backbone
-
Vector backbonepCAMBIA-0380
-
Backbone manufacturerCAMBIA
- Backbone size w/o insert (bp) 6812
- Total vector size (bp) 10264
-
Modifications to backboneThe hygromycin gene in ternary vector pC-HYG-YR (Addgene plasmid# 61765) was replaced with the bar gene to construct pC-BAR-YR.
-
Vector typeTernary vector for Agrobacterium mediated transformation of fungi
-
Selectable markersURA3, Basta
Growth in Bacteria
-
Bacterial Resistance(s)Kanamycin, 50 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert 1
-
Gene/Insert nameBar gene
-
Alt namebiaphalos/BASTA resistance gene
-
SpeciesStreptomyces hygroscopicus
-
Insert Size (bp)552
-
GenBank ID
- Promoter trpC promoter
Cloning Information for Gene/Insert 1
- Cloning method Restriction Enzyme
- 5′ cloning site EcoRI (not destroyed)
- 3′ cloning site HindIII (not destroyed)
- 5′ sequencing primer LB-F1 (5'-gtggtgtaaacaaattgacgc-3')
- 3′ sequencing primer RB-F1 (5'-ggataaaccttttcacgccc-3') (Common Sequencing Primers)
Gene/Insert 2
-
Gene/Insert name2 micron origin or replication and URA3 gene for S. cerevisiae
-
SpeciesS. cerevisiae (budding yeast)
-
Insert Size (bp)2487
-
GenBank IDKC562906.1
Cloning Information for Gene/Insert 2
- Cloning method Restriction Enzyme
- 5′ cloning site SacII (not destroyed)
- 3′ cloning site SacII (not destroyed)
- 5′ sequencing primer ccgcggTTAGTTTTGCTGGCCGCATC
- 3′ sequencing primer ccgcggCGCTTCCACAAACATTGCTC (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byBar gene was cloned from pCAMB-BAR (Kramer et al 2009)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Jason Rudd, (Rothamsted research UK) provided Agrobacterium tumfaciens vector pCAMB-BAR (pCAMBIA-0380 backbone with bar gene under Neurospora crassa trpC promoter) For more information see: Kramer et al.. Fungal Genetics and Biology, 46, 667-681. https://www.ncbi.nlm.nih.gov/pubmed/19520179.
Bar gene from pCAMB-BAR was recombined into pC-HYG-YR (Addgene plasmid# 61765) through yeast recombineering to replace hygromycin gene and create pC-BAR-YR.
Restriction sites EcoRI and HindIII can be used to linearise the vector inside left (LB) and right (RB) borders of the TDNA. Note that the trpC promoter and bar sequences are present within the EcoRI and HindII cloning sites.
See Supplementary Fig. S1. of associated publication for schematic of single step assembly of gene deletion ternary vectors using yeast recombinational cloning.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pC-BAR-YR was a gift from Ken Haynes (Addgene plasmid # 61766 ; http://n2t.net/addgene:61766 ; RRID:Addgene_61766) -
For your References section:
Exploitation of sulfonylurea resistance marker and non-homologous end joining mutants for functional analysis in Zymoseptoria tritici. Sidhu YS, Cairns TC, Chaudhari YK, Usher J, Talbot NJ, Studholme DJ, Csukai M, Haynes K. Fungal Genet Biol. 2015 Jun;79:102-9. doi: 10.1016/j.fgb.2015.04.015. 10.1016/j.fgb.2015.04.015 PubMed 26092796