Phosphoinositide Biosensors Kit
(Kit #
1000000261
)
Depositing Lab: Hannes Maib, David Murray
Membrane identity is key to cellular function and partly determined by the phosphoinositides, a small family of phospholipids with differentially phosphorylated inositol headgroups. We have developed a suite of recombinant biosensors to detect the whole family of phosphoinositides in fixed cells and tissues. This approach bypasses the need to overexpress these probes and is compatible with multiplex and super resolution microscopy.
This kit will be sent as individual bacterial stabs at room temperature.
Original Publication
Recombinant biosensors for multiplex and super-resolution imaging of phosphoinositides. Maib H, Adarska P, Hunton R, Vines JH, Strutt D, Bottanelli F, Murray DH. J Cell Biol. 2024 Jun 3;223(6):e202310095. doi: 10.1083/jcb.202310095. Epub 2024 Apr 5. PubMed (Link opens in a new window) Article (Link opens in a new window)
Description
The most common way to visualize the subcellular localization of phosphoinositides is to ectopically overexpress the lipid effectors fused to fluorescent proteins as biosensors (Wills et al., 2018). This approach allows for the detection of membrane identity changes in live cells and has been of immense value over the last decades. However, it also suffers from a major drawback: as these biosensors are generated to have a high affinity, they can all potentially out-compete endogenous effector proteins and thereby perturb the very pathway that is under investigation. In contrast, biosensors with lower affinity and expression levels are themselves out-competed by endogenous effectors. Furthermore, this overexpression-based approach is limited by the ability to deliver DNA into cells or requires time and labor-extensive genomic engineering. It is therefore still a challenge for experimentalists to visualize these critically important lipids, especially in more complex systems such as 3D cell cultures or whole tissues.
One way to circumvent these pitfalls has been to stain for phosphoinositides after fixation and permeabilization using immunocytochemical techniques based on recombinant biosensors (Gillooly et al., 2000; Watt et al., 2002) and antibodies (Hammond et al., 2009; Maekawa and Fairn, 2014; Marat et al., 2017). While this approach is unable to provide dynamic information, it does avoid perturbation of phosphoinositide signaling and does not require overexpression. However, the need for fixatives and detergents requires careful optimizations to avoid artifacts, and the available antibodies require different staining protocols for the visualization of distinct pools of their respective targets (Hammond et al., 2009). As such, there is currently no reliable tool to comprehensively visualize all subcellular phosphoinositide pools with a unifying staining approach.
To address these shortcomings, we have developed recombinant biosensors against all eight phosphoinositides in combination with self-labeling protein tags (SNAP). With the exception of the only known biosensor against PI(5)P, all of these probes are easy to purify using standard bacterial expression systems and demonstrate excellent specificities, as determined using an in vitro supported lipid bilayer approach. Using a single, unified staining protocol, we show that these probes reliably visualize at least six out of the eight phosphoinositide species after fixation and permeabilization. We successfully reproduce the known subcellular localizations of these phosphoinositides and verify the specificity of the staining approach using a range of lipid kinase inhibitors. With the exception of PI(5)P, and with some caveats in detecting PI, this toolkit enables the reliable and reproducible detection of PI(3)P, PI(4)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3.
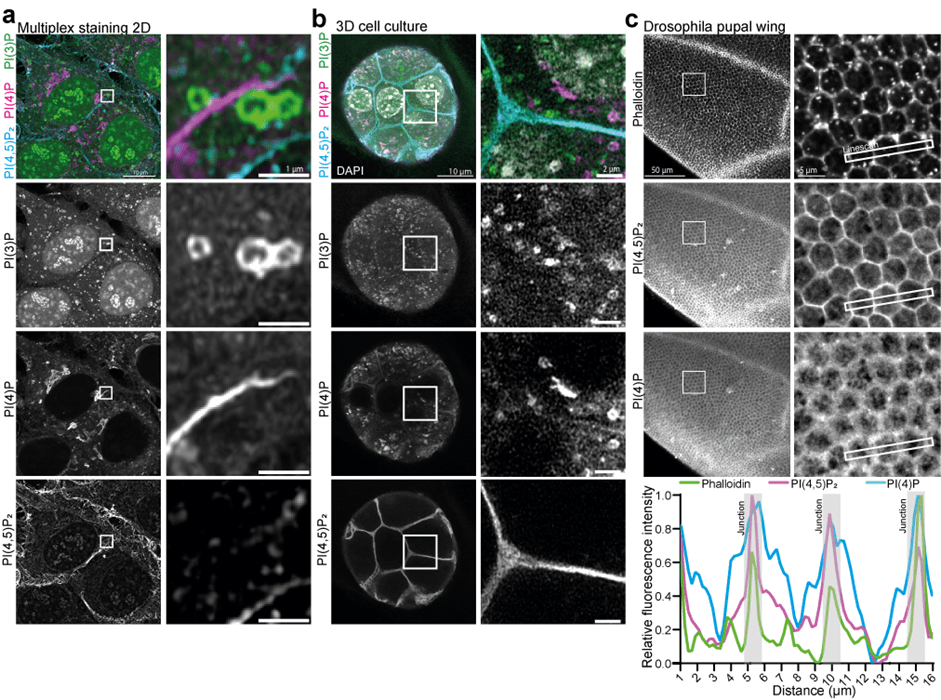
Figure 1: Multiplex staining of phosphoinositides across scales. (a) HeLa cells were fixed, permeabilized, and stained using recombinant biosensors against PI(3)P, PI(4)P, and PI(4,5)P2, conjugated respectively to Alexa488, 546, and 647. (b) NMuMG spheroids were grown in Matrigel and stained with the same combination. (c) Drosophila pupal wings were dissected and stained with the PI(4,5)P2 and PI(4)P biosensors conjugated to Alexa647 and 546, together with Phalloidin conjugated to Alexa488, to visualize the actin cytoskeleton and cellular junctions.
References
Wills, R. C., Goulden, B. D., & Hammond, G. R. V. (2018). Genetically encoded lipid biosensors. Mol Biol Cell, 29(13), 1526–1532. https://doi.org/10.1091/mbc.E17-12-0738 (Link opens in a new window) PMID: 29953345 (Link opens in a new window)
Gillooly, D. J., Morrow, I. C., Lindsay, M., Gould, R., Bryant, N. J., Gaullier, J. M., Parton, R. G., & Stenmark, H. (2000). Localization of phosphatidylinositol 3 phosphate in yeast and mammalian cells. EMBO J, 19(17), 4577–4588. https://doi.org/10.1093/emboj/19.17.4577 (Link opens in a new window) PMID: 10970851 (Link opens in a new window)
Watt M. J., Heigenhauser G. J. F., Dyck D. J., & Spriet L. L. (2002). Intramuscular triacylglycerol, glycogen and acetyl group metabolism during 4 h of moderate exercise in man.. J Physiol, 541(Pt 3), 969–978. https://doi.org/10.1113/jphysiol.2002.018820 (Link opens in a new window) PMID: 12068055 (Link opens in a new window)
Hammond, G. R. V., Schiavo, G., & Irvine, R. F. (2009). Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2). Biochem J, 422(1), 23–35. https://doi.org/10.1042/BJ20090428 (Link opens in a new window) PMID: 19508231 (Link opens in a new window)
Hammond, G. R. V., Sim, Y., Lagnado, L., & Irvine, R. F. (2009). Reversible binding and rapid diffusion of proteins in complex with inositol lipids serves to coordinate free movement with spatial information. J Cell Biol, 184(2), 297–308. https://doi.org/10.1083/jcb.200809073 (Link opens in a new window) PMID: 19153221 (Link opens in a new window)
Maekawa M. & Fairn G. D. (2014). Molecular probes to visualize the location, organization and dynamics of lipids. J Cell Sci., 127(Pt 22), 4801-12. https://doi.org/10.1242/jcs.150524 (Link opens in a new window) PMID: 25179600 (Link opens in a new window)
Marat, A. L., Wallroth, A., Lo, W. T., Müller, R., Norata, G. D., Falasca, M., Schultz, C., & Haucke, V. (2017). mTORC1 activity repression by late endosomal phosphatidylinositol 3,4 bisphosphate. Science, 356(6341), 968–972. https://doi.org/10.1126/science.aaf8310 (Link opens in a new window) PMID: 28572395 (Link opens in a new window)
Kit Documentation
A detailed protocol is provided in the Materials and Methods section of the publication.
How to Cite this Kit
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which they were created, and include Addgene in the Materials and Methods of your future publications.
For your Materials and Methods section:
"The Phosphoinositide Biosensors Kit was a gift from David Murray (Addgene kit #1000000261)."
For your Reference section:
Recombinant biosensors for multiplex and super-resolution imaging of phosphoinositides. Maib H, Adarska P, Hunton R, Vines JH, Strutt D, Bottanelli F, Murray DH. J Cell Biol. 2024 Jun 3;223(6):e202310095. doi: 10.1083/jcb.202310095. Epub 2024 Apr 5. PubMed (Link opens in a new window) Article (Link opens in a new window)
The Phosphoinositide Biosensors Kit contains 6 plasmids. Please refer to the individual plasmid pages below for more details on each plasmid in this kit:


