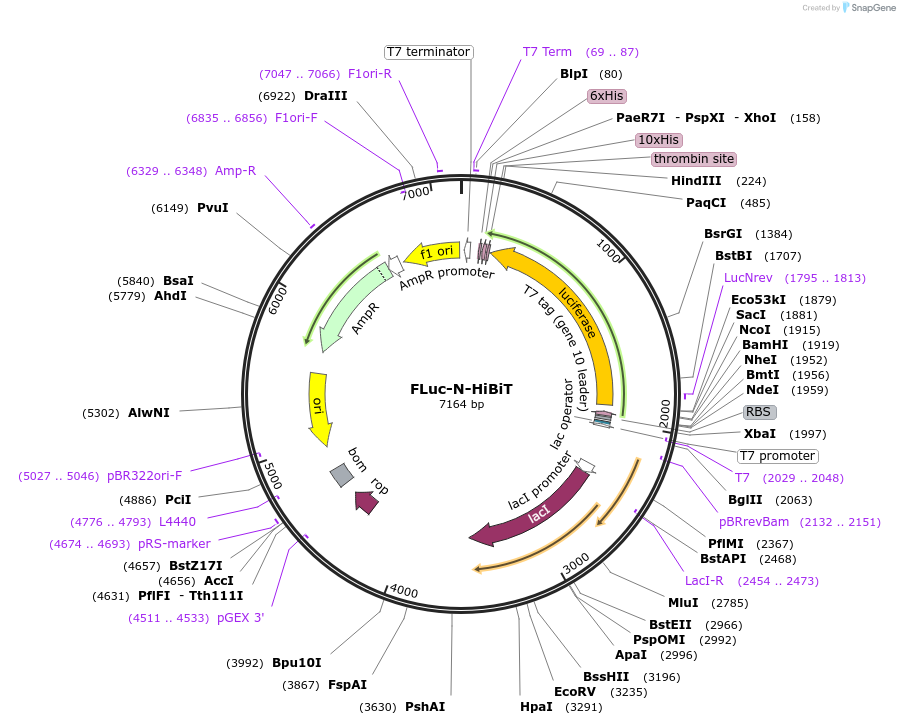
FLuc-N-HiBiT
(Plasmid
#207117)
-
PurposeBacterially expressed HiBiT-FLuc for Ni-NTA purification
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 207117 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepET21a (+)
-
Backbone manufacturerMilliporeSigma (Novagen)
- Backbone size w/o insert (bp) 5370
- Total vector size (bp) 7164
-
Vector typeBacterial Expression
-
Selectable markersN/A
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberLow Copy
Gene/Insert
-
Gene/Insert nameluciferin 4-monooxygenase
-
Alt nameFLuc_N_HiBiT
-
Alt nameHiBiT-FLuc
-
Alt namePhotinus pyralis luciferase
-
SpeciesP. pyralis
-
Insert Size (bp)1760
-
MutationCodon optmization for expression in BL21DE3
-
GenBank IDM15077.1
- Promoter T7 Promotor
-
Tags
/ Fusion Proteins
- HiBiT Tag (N terminal on insert)
- His-tag with thrombin site (C terminal on insert)
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site BamH1 (unknown if destroyed)
- 3′ cloning site Hind3 (unknown if destroyed)
- 5′ sequencing primer T7 promotor primer #69348-3
- 3′ sequencing primer T7 terminator primer #69337-3
- (Common Sequencing Primers)
Resource Information
-
A portion of this plasmid was derived from a plasmid made bysynthesized at BIO Basic, Inc. for us.
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
FLuc-N-HiBiT was a gift from James Inglese (Addgene plasmid # 207117 ; http://n2t.net/addgene:207117 ; RRID:Addgene_207117) -
For your References section:
A general assay platform to study protein pharmacology using ligand-dependent structural dynamics. Ciulla DA, Dranchak PK, Aitha M, van Neer RHP, Shah D, Tharakan R, Wilson KM, Wang Y, Braisted JC, Inglese J. Nat Commun. 2025 May 10;16(1):4342. doi: 10.1038/s41467-025-59658-6. 10.1038/s41467-025-59658-6 PubMed 40346061