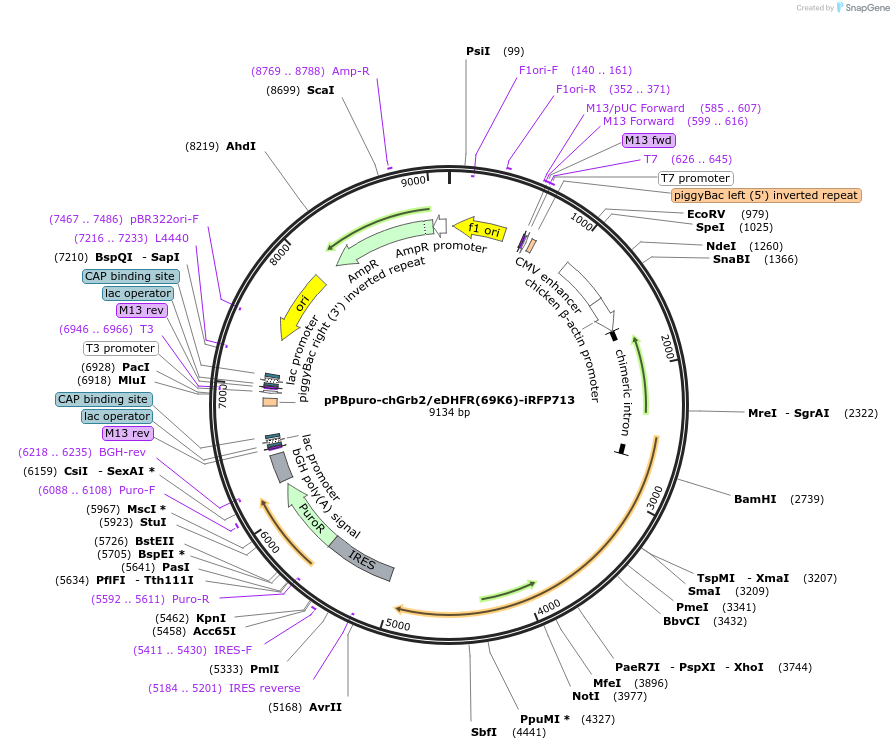
pPBpuro-chGrb2/eDHFR(69K6)-iRFP713
(Plasmid
#214828)
-
PurposeEncoding Grb2-eDHFR(69K6) chimera fused to iRFP713
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 214828 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepPB
- Backbone size w/o insert (bp) 6915
- Total vector size (bp) 9126
-
Vector typeMammalian Expression
-
Selectable markersPuromycin
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)NEB Stable
-
Copy numberUnknown
Gene/Insert
-
Gene/Insert nameGrb2(1-59)-eDHFR(69K6)-Grb2(152-216)-iRFP713
-
SpeciesH. sapiens (human)
-
Insert Size (bp)2211
- Promoter CAG promoter
-
Tag
/ Fusion Protein
- iRFP713 (C terminal on insert)
Cloning Information
- Cloning method Ligation Independent Cloning
- 5′ sequencing primer none
- 3′ sequencing primer none
- (Common Sequencing Primers)
Resource Information
-
A portion of this plasmid was derived from a plasmid made byDr. Shinya Tsukiji (Addgene plasmid # 178854: pCAGGS-chGrb2/eDHFR(69K6)-iRFP713
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pPBpuro-chGrb2/eDHFR(69K6)-iRFP713 was a gift from Michiyuki Matsuda (Addgene plasmid # 214828 ; http://n2t.net/addgene:214828 ; RRID:Addgene_214828) -
For your References section:
Front-biased activation of Ras-Rab5-Rac1 loop coordinates collective cell migration. Jikko Y, Deguchi E, Matsuda K, Hino N, Tsukiji S, Matsuda M, Terai K. J Cell Sci. 2025 Jul 16:jcs.263779. doi: 10.1242/jcs.263779. 10.1242/jcs.263779 PubMed 40667649