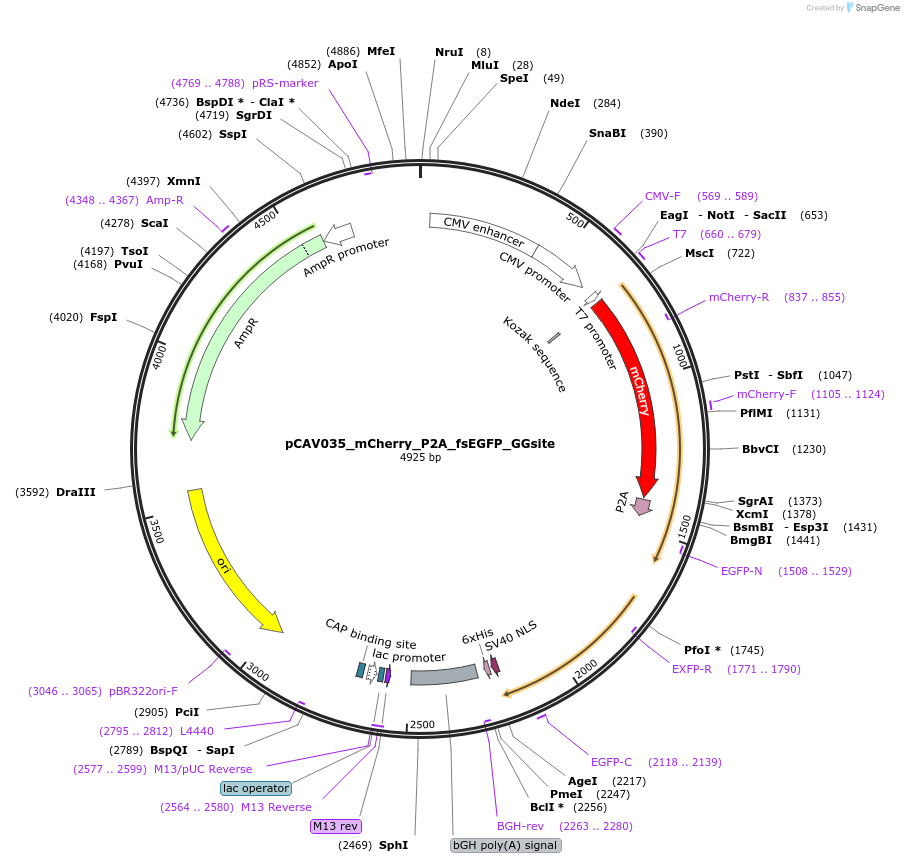
pCAV035_mCherry_P2A_fsEGFP_GGsite
(Plasmid
#219807)
-
PurposeFluorescent Reporter for MUTYH-Specific Activity; Containing BsaI Recognition Sequence at Frameshifted EGFP; For OG:A Oligo Insertion at Golden Gate (GG) Site at Codon 34
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 219807 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepCAG
- Backbone size w/o insert (bp) 3435
- Total vector size (bp) 4923
-
Modifications to backboneThe EGFP portion of this plasmid is frameshifted, which makes it non-fluorescent, but contains a golden gate site for BsaI recognition. Once digested with BsaI, an OG-A lesion can be ligated in at codon 34 of EGFP making it an assay for MUTYH-specific activity.
-
Vector typeMammalian Expression
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert namemCherry
-
SpeciesSynthetic
-
GenBank IDAY678264.1
- Promoter CMV
-
Tag
/ Fusion Protein
- mCherry (N terminal on backbone)
Cloning Information
- Cloning method Other
- 5′ sequencing primer agctactaacttcagcctgctgaagc
- 3′ sequencing primer caggaaacagctatgac
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pCAV035_mCherry_P2A_fsEGFP_GGsite was a gift from Alexis Komor (Addgene plasmid # 219807 ; http://n2t.net/addgene:219807 ; RRID:Addgene_219807) -
For your References section:
Precision genome editing and in-cell measurements of oxidative DNA damage repair enable functional and mechanistic characterization of cancer-associated MUTYH variants. Vasquez CA, Osgood NRB, Zepeda MU, Sandel DK, Cowan QT, Peiris MN, Donoghue DJ, Komor AC. Nucleic Acids Res. 2025 Mar 20;53(6):gkaf037. doi: 10.1093/nar/gkaf037. 10.1093/nar/gkaf037 PubMed 40156857