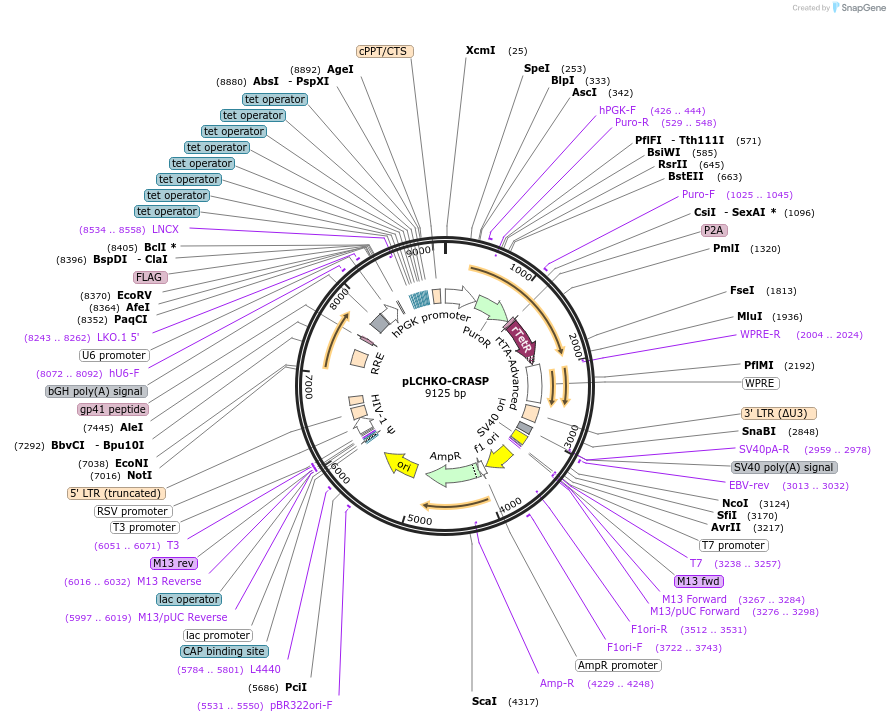
pLCHKO-CRASP
(Plasmid
#231928)
-
Purpose(Empty Backbone) Empty backbone for CRASP-Seq experiments
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 231928 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepLCHKOv3 (Addgene Plasmid #209025)
-
Vector typeLentiviral
-
Selectable markersPuromycin
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature30°C
-
Growth Strain(s)NEB Stable
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameNone
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Please visit https://doi.org/10.1101/2025.04.25.648738 for bioRxiv preprint.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pLCHKO-CRASP was a gift from Thomas Gonatopoulos-Pournatzis (Addgene plasmid # 231928 ; http://n2t.net/addgene:231928 ; RRID:Addgene_231928) -
For your References section:
RNA-coupled CRISPR screens reveal ZNF207 as a regulator of LMNA aberrant splicing in progeria. Behera AK, Kim JJ, Kordale S, Pekovic F, Damodaran AP, Kumari B, Vidak S, Dickson E, Xiao MS, Duncan G, Andresson T, Misteli T, Valkov E, Gonatopoulos-Pournatzis T. Mol Cell. 2026 Jan 8;86(1):41-59.e15. doi: 10.1016/j.molcel.2025.12.003. Epub 2025 Dec 30. 10.1016/j.molcel.2025.12.003 PubMed 41475346