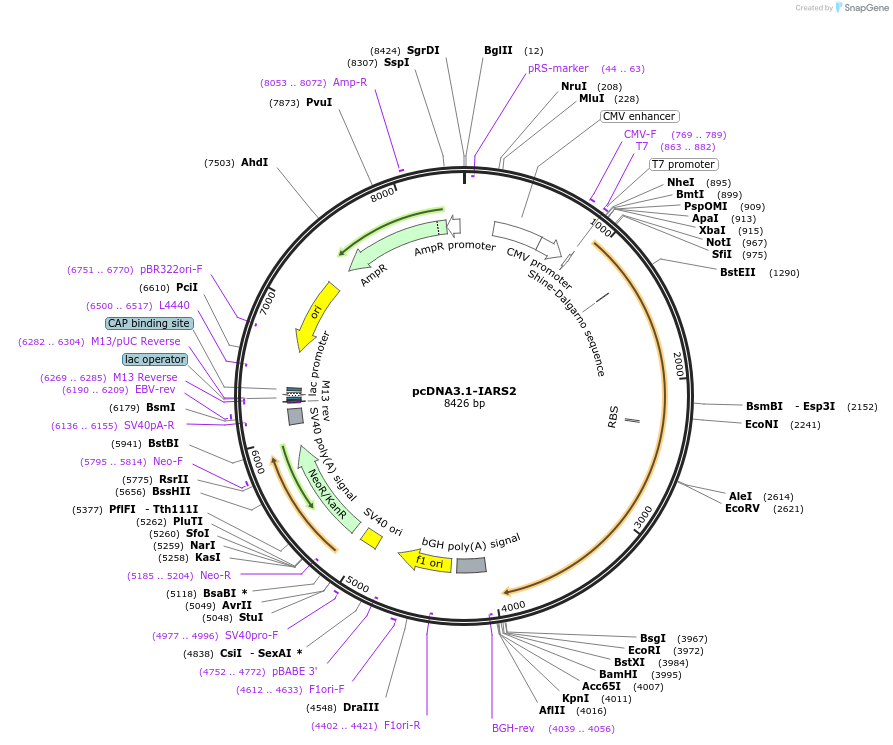
pcDNA3.1-IARS2
(Plasmid
#232337)
-
PurposeExpresses human IARS2 (ENST00000366922.3, NM_018060.4) in mammalian cells
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 232337 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepcDNA3.1 (-)
-
Backbone manufacturerInvitrogen (Thermo Fisher)
- Backbone size w/o insert (bp) 5427
- Total vector size (bp) 8445
-
Vector typeMammalian Expression
-
Selectable markersNeomycin (select with G418)
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameIARS2
-
Alt nameENST00000366922.3
-
SpeciesH. sapiens (human)
-
GenBank IDNM_018060.4
-
Entrez GeneIARS2 (a.k.a. CAGSSS, ILERS)
- Promoter CMV
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site XhoI (destroyed during cloning)
- 3′ cloning site EcoRI (not destroyed)
- 5′ sequencing primer CMV-F: CGCAAATGGGCGGTAGGCGTG
- 3′ sequencing primer BGH-R: TAGAAGGCACAGTCGAGG
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byYuxin Bi (毕雨馨 in simplified Chinese), the first author of the article, constructed this plasmid during her master's degree. She graduated and become a PHD candidate in other institute for now.
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Note1:
At the 5' of the IARS2 insert, we use diffrernt restriction enzyme for the insert and the backbone. Xho I for pcDNA3.1 (-) and Sal I for IARS2 ORF. The Xho I and Sal I share a compatible cohesive end. Ligation between the compatible cohesive ends generated by different enzymes destroy the 5' restriction site. We use this cloning method because that IARS2 ORF (ENST00000366922.3) bears intrinsic Xho I site.
Note2:
The IARS2 ORF is long, so we used additional primers (5'->3') for sanger sequencing as follows:
pc3.1-IARS2-P1: tgggaagtatgaagccaaacagt
pc3.1-IARS2-P2: gaccatggcaaaaggaacgg
pc3.1-IARS2-P3: caacagccaaaccactgagc
pc3.1-IARS2-P4: tgctcaatgctgccagaga
Note3:
The putative Kozak sequence (ENST00000366922.3:c.-6~-1) was also included
Note4:
There is a discrepancy in the backbone in the SV40 promoter where the Addgene NGS sequence contains an indel compared to the depositor reference. However, this indel does not affect the function of the plasmid because the SV40 promoted G418 resistant gene was not used in the study. The depositor used these plasmids for transient overexpression, not stable expression.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pcDNA3.1-IARS2 was a gift from Quanfu Ma (Addgene plasmid # 232337 ; http://n2t.net/addgene:232337 ; RRID:Addgene_232337)