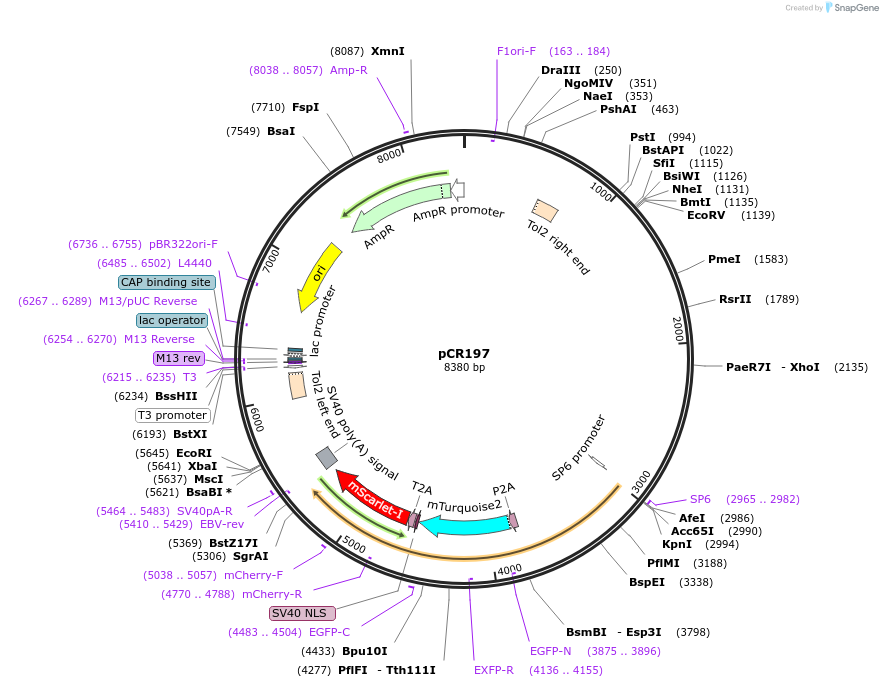
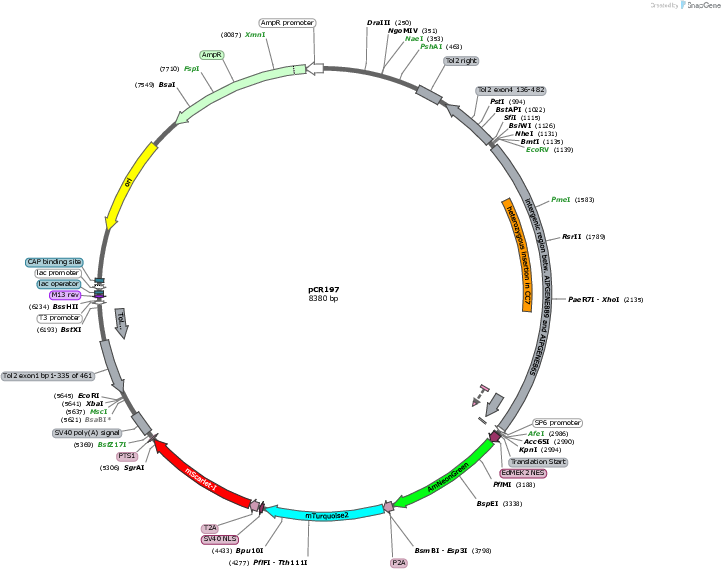
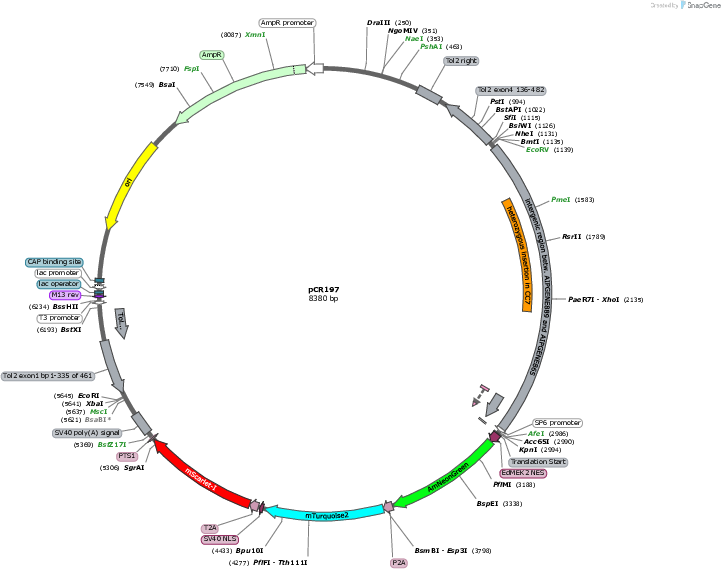
pCR197
(Plasmid
#240643)
-
PurposeExpression of the tri-cistronic construct NES-AmNeonGreen-P2A-mTurquoise2-NLS-T2A-mScarlet-I-PTS1 in Exaiptasia diaphana; In vitro transcription of the same construct via SP6 polymerase
-
Depositing Lab
-
Publication
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 240643 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepT2HE
-
Backbone manufacturerTim Howes
- Backbone size w/o insert (bp) 3854
- Total vector size (bp) 6440
-
Vector typeGene expression and genomic integration in fish
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert 1
-
Gene/Insert nameAIPGENE865 promoter
-
Alt nameExaiptasia diaphana Elongation Factor 1 alpha promoter
-
SpeciesExaiptasia diaphana
-
Insert Size (bp)1810
-
Mutationnaturally occurring 749-bp insertion at bp 628 (from bp 1 of plasmid map), corrected exon/intron positions (in agreement with XM_021048156.2).
-
GenBank IDKXJ12416.1 XM_021048156.2
Cloning Information for Gene/Insert 1
- Cloning method Gibson Cloning
- 5′ sequencing primer Tol2right_up_seq_fwd (CTGTTCAGACACCCATATCC), P1_fwd (CGTACGCTAGCGATATCGG)
- 3′ sequencing primer P2_2019_rev (AGCCATTATGGTACCAGCGC)
- (Common Sequencing Primers)
Gene/Insert 2
-
Gene/Insert nameSP6 promoter
-
Insert Size (bp)19
-
GenBank ID
Cloning Information for Gene/Insert 2
- Cloning method Gibson Cloning
- 5′ sequencing primer AmNG_mid_seq_rev (GACCGTCAGCAGGGAAACC)
- (Common Sequencing Primers)
Gene/Insert 3
-
Gene/Insert nameAmNeonGreen
-
SpeciesSynthetic
-
Insert Size (bp)708
-
MutationCodon-optimized for Exaiptasia diaphana, amino acid sequence identical to original mNeonGreen.
-
GenBank IDKC295282.1
-
Tags
/ Fusion Proteins
- NES (from Exaiptasia diaphana MEK2) (N terminal on insert)
- P2A site (C terminal on insert)
Cloning Information for Gene/Insert 3
- Cloning method Gibson Cloning
- 5′ sequencing primer AmNG_mid_seq_rev (GACCGTCAGCAGGGAAACC)
- 3′ sequencing primer P2A_seq_rev (AGGACCGGGGTTTTCTTCC)
- (Common Sequencing Primers)
Gene/Insert 4
-
Gene/Insert namemTurquoise2
-
SpeciesSynthetic
-
Insert Size (bp)717
-
GenBank IDKM670033.1
-
Tags
/ Fusion Proteins
- NLS (from SV40 T antigen) (C terminal on insert)
- T2A site (C terminal on insert)
Cloning Information for Gene/Insert 4
- Cloning method Gibson Cloning
- 5′ sequencing primer T2A_seq_rev (GGGGCCGGGATTTTCCTCC)
- 3′ sequencing primer RFPs_mid_seq_rev (GTCTTCTTCTGCATTACGGGG)
- (Common Sequencing Primers)
Gene/Insert 5
-
Gene/Insert namemScarlet-I
-
SpeciesSynthetic
-
Insert Size (bp)696
-
GenBank IDKY021424.1
-
Tag
/ Fusion Protein
- Peroxisomal targeting site 1 (C terminal on insert)
Cloning Information for Gene/Insert 5
- Cloning method Gibson Cloning
- 5′ sequencing primer RFPs_mid_seq_rev (GTCTTCTTCTGCATTACGGGG)
- 3′ sequencing primer RFPs_mid_seq_fwd (GAGCGCGTGATGAACTTCG)
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made bypT2HE (backbone): O’Brown NM, Summers BR, Jones FC, Brady SD, Kingsley DM. 2015. A recurrent regulatory change underlying altered expression and Wnt response of the stickleback armor plates gene EDA. eLife. 4:e05290. doi:10.7554/eLife.05290. mNeonGreen: Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al. 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 10(5):407–409. doi:10.1038/nmeth.2413. mTurquoise2: Goedhart J, Von Stetten D, Noirclerc-Savoye M, Lelimousin M, Joosen L, Hink MA, Van Weeren L, Gadella TWJ, Royant A. 2012. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat Commun. 3(1):751. doi:10.1038/ncomms1738. mScarlet-I: Bindels DS, Haarbosch L, Van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, et al. 2017. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods. 14(1):53–56. doi:10.1038/nmeth.4074. Plasmid mScarlet-I-mTurquoise2 (Addgene plasmid # 98839): Mastop M, Bindels DS, Shaner NC, Postma M, Gadella TWJ, Goedhart J. 2017. Characterization of a spectrally diverse set of fluorescent proteins as FRET acceptors for mTurquoise2. Sci Rep. 7(1):11999. doi:10.1038/s41598-017-12212-x.
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
The whole intergenic region between AIPGENE865 (KXJ12416.1; Elongation Factor 1alpha) and AIPGENE899 was cloned as promoter fragment. The original gene annotation for AIPGENE865 is incorrect, the automatic NCBI annotation is correct in regards of exons, introns and translation start site. We found that Aiptasia strain CC7 was actually heterozygous for that locus: one allele was mostly in agreement with the genome sources but the other one had a couple of SNPs and a large, 749-bp insert in the intergenic region. The longer version with insert was used as promoter for this plasmid.
The Aiptasia MEK2 locus was found to be erroneously split into AIPGENES3280 & 23794 on two different scaffolds due to an assembly gap in the Baumgarten et al., 2015 genome. Correct CDS was identified from sequencing based on cDNA of strain CC7.
AmNeonGreen is a version of mNeonGreen codon-optimized for Aiptasia (based on the transcriptome of Lehnert et al., 2014) with the identical protein sequence.
Baumgarten S, Simakov O, Esherick LY, Liew YJ, Lehnert EM, Michell CT, Li Y, Hambleton EA, Guse A, Oates ME, et al. 2015. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci. 112(38):11893–11898. doi:10.1073/pnas.1513318112.
Lehnert EM, Mouchka ME, Burriesci MS, Gallo ND, Schwarz JA, Pringle JR. 2014. Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3 Bethesda Md. 4(2):277–295. doi:10.1534/g3.113.009084.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pCR197 was a gift from John Pringle (Addgene plasmid # 240643 ; http://n2t.net/addgene:240643 ; RRID:Addgene_240643)