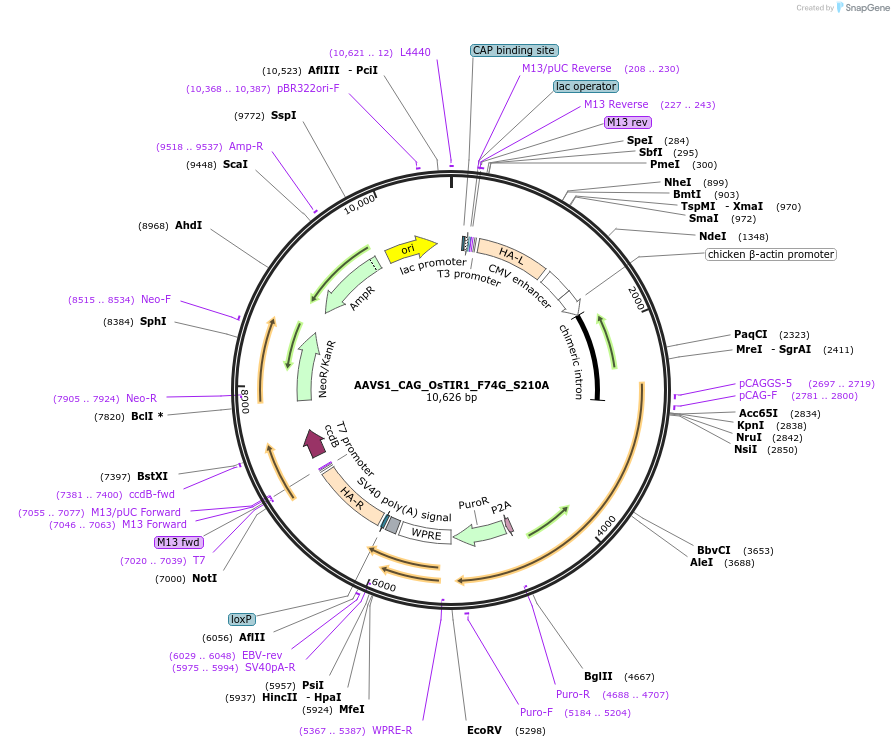
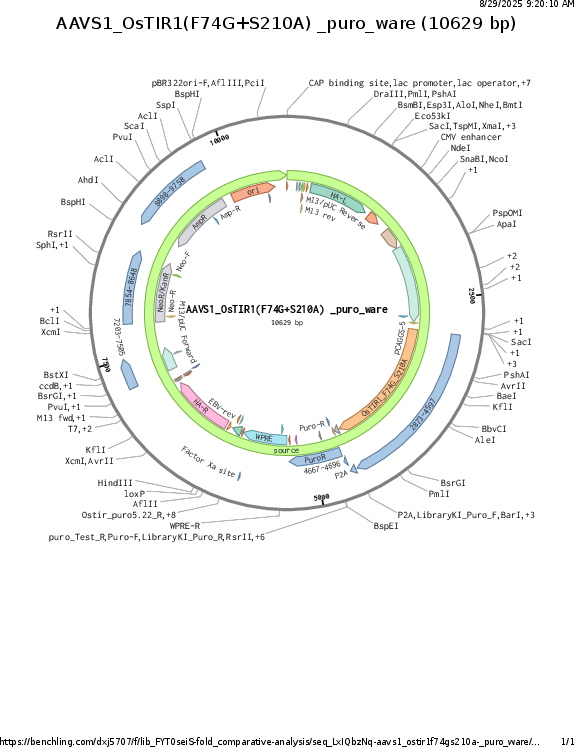
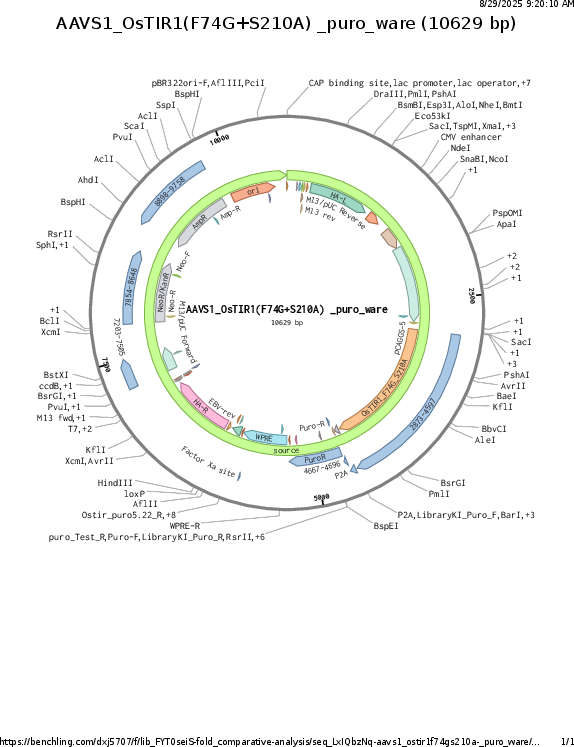
AAVS1_CAG_OsTIR1_F74G_S210A
(Plasmid
#242189)
-
PurposeAID 2.0's higher degradation efficiency came with target-specific basal degradation and slower recovery rates. To address these limitations, we employed base-editing-mediated mutagenesis followed by s
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 242189 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepMK381
-
Vector typeMammalian Expression
-
Selectable markersPuromycin
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameCAG_OsTIR1_F74G_S210A
-
SpeciesOryza sativa (rice)
-
MutationS210A
-
GenBank ID4335696
- Promoter CAG
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site NdeI (not destroyed)
- 3′ cloning site MfeI (not destroyed)
- 5′ sequencing primer gcaacgtgctggttattgtg
- (Common Sequencing Primers)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
AAVS1_CAG_OsTIR1_F74G_S210A was a gift from Mazhar Adli (Addgene plasmid # 242189 ; http://n2t.net/addgene:242189 ; RRID:Addgene_242189) -
For your References section:
Systematic comparison and base-editing-mediated directed protein evolution and functional screening yield superior auxin-inducible degron technology. Xing D, Bai T, Neyisci O, Paylakhi SZ, Duval AJ, Tekin Y, Adli M. Nat Commun. 2025 Jul 18;16(1):6631. doi: 10.1038/s41467-025-61848-1. 10.1038/s41467-025-61848-1 PubMed 40681502