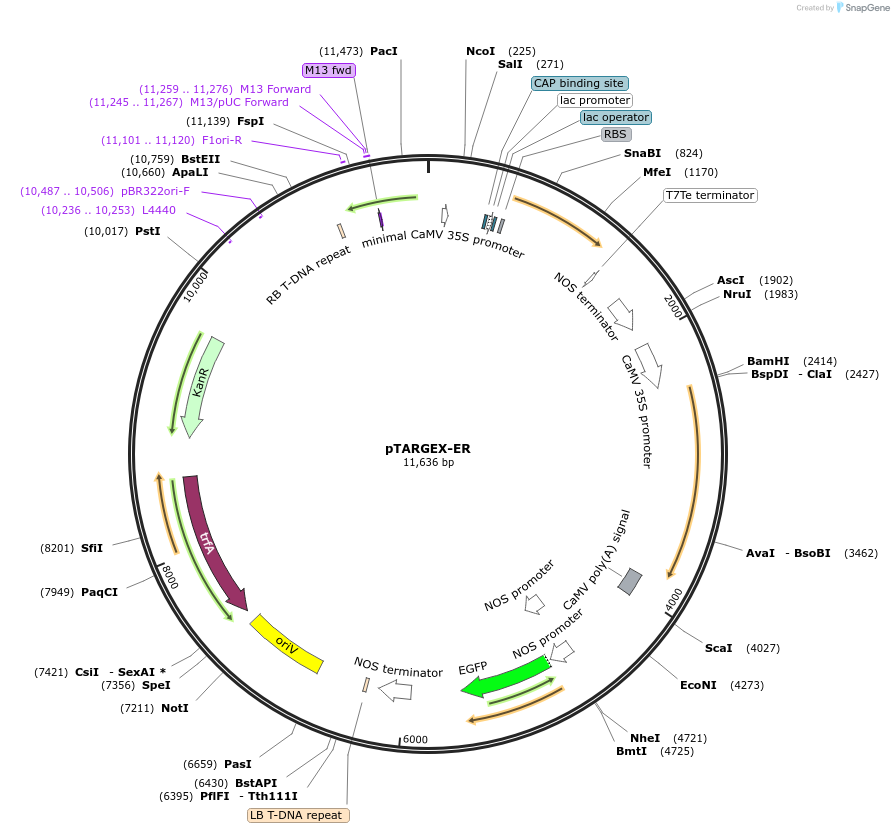
pTARGEX-ER
(Plasmid
#242449)
-
Purpose(Empty Backbone) Targets high levels of recombinant protein for retention in ER. Empty vector. Clone via SapI restriction site. mCherry dropout cassette will enable Red/White screening in E.coli.
-
Depositing Lab
-
Publication
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 242449 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepTARGEX
-
Backbone manufactureruOttawa iGEM team 2024
- Backbone size (bp) 11636
-
Modifications to backboneAddition of N-terminal signal peptide derived from the Nicotiana tabacum pathogenesis-related protein 1b (PR1b) and a canonical C-terminal ER-retrieval motif (KDEL)
-
Vector typePlant Expression
- Promoter double 35S
-
Tags
/ Fusion Proteins
- Signal peptide derived from the Nicotiana tabacum pathogenesis-related protein 1b (N terminal on backbone)
- ER-retrieval motif (KDEL) (C terminal on backbone)
Growth in Bacteria
-
Bacterial Resistance(s)Kanamycin, 50 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Growth instructionsClone and maintain in E. coli. For expression in planta, transform into suitable Agrobacterium tumefaciens strain and grow at 28C.
-
Copy numberLow Copy
Cloning Information
- Cloning method Restriction Enzyme
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pTARGEX-ER was a gift from Allyson MacLean (Addgene plasmid # 242449 ; http://n2t.net/addgene:242449 ; RRID:Addgene_242449) -
For your References section:
pTARGEX Vectors: A Versatile Toolbox for Plant-Based Protein Expression. Chartrand-Pleau C, Gallipeau-Burns E, Jain A, Boddy V, Fakhouri P, Rehmani S, Thomas T, Drapeau M, VanderBurgt JT, Boudigou A, Damry AM, MacLean AM. Front. Synth. Biol. 3:1619871 10.3389/fsybi.2025.1619871