Promoters
A promoter is a region of DNA where transcription of a gene is initiated. Promoters are a vital component of expression vectors because they control the binding of the RNA polymerase to DNA. RNA polymerase transcribes DNA to mRNA, which is ultimately translated into a functional protein. Thus, the promoter region controls when and where in the organism your gene of interest is expressed.
Promoters are about 100–1,000 base pairs long and are adjacent and typically upstream (5’) of the sense or coding strand of the transcribed gene (Figure 1). The coding strand is the DNA strand that encodes codons and whose sequence corresponds to the mRNA transcript produced. The antisense strand is referred to as the template strand or noncoding strand, as this is the strand that is transcribed by the RNA polymerase.
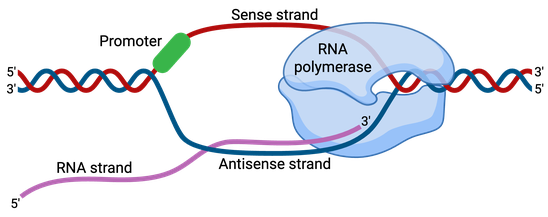
DNA sequences called response elements are located within promoter regions, and they provide a stable binding site for the RNA polymerase and transcription factors. Transcription factors are proteins which recruit RNA polymerase and control and regulate the transcription of DNA into mRNA.
Promoter binding is very different in bacteria compared to eukaryotes. In bacteria, the core RNA polymerase requires an associated sigma factor for promoter recognition and binding. On the other hand, the process in eukaryotes is much more complex. Eukaryotes require a minimum of seven transcription factors in order for RNA polymerase II (a eukaryote-specific RNA polymerase) to bind to a promoter. Transcription is tightly controlled in both bacteria and eukaryotes by various DNA regulatory sequences, including enhancers, boundary elements, insulators, and silencers.
Bacterial Promoters
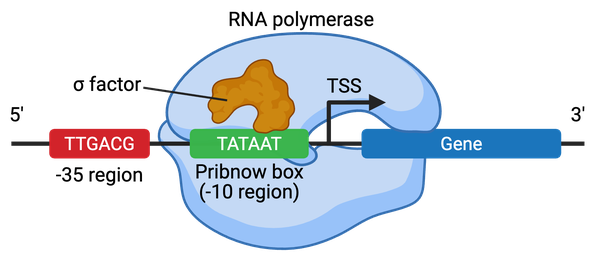
Promoters in bacteria contain two short DNA sequences located at the -10 (10 bp 5' or upstream) and -35 positions from the transcription start site (TSS) (Figure 2). The Pribnow box (TATAAT) is located at the -10 position and is essential for transcription initiation. The -35 position, simply titled the -35 element, typically consists of the sequence TTGACA, and this element controls the rate of transcription. Bacterial cells contain sigma factors that assist the RNA polymerase in binding to the promoter region. Each sigma factor recognizes different core promoter sequences.
Operons
Although bacterial transcription is simpler than eukaryotic transcription, bacteria still have complex systems of gene regulation, like operons. Operons are a cluster of different genes that are controlled by a single promoter and operator. Operons are common in prokayotes, specifically bacteria, but have also been discovered in eukaryotes. Operons consist of a promoter (which is recognized by the RNA polymerase), an operator (a segment of DNA in which a repressor or activator can bind), and the structural genes that are transcribed together.
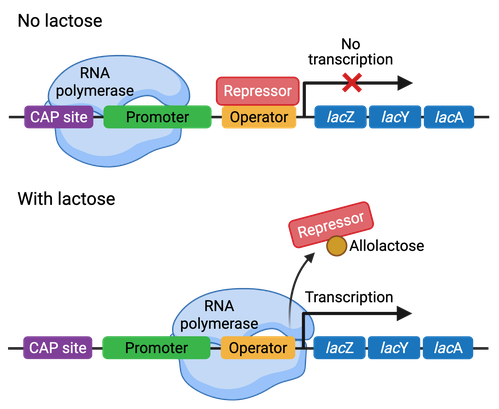
Operon regulation can be either negative or positive. Repressor proteins bind to negative-repressible operons and prevent transcription. When an inducer molecule binds to the repressor, it changes its conformation, preventing its binding to the operator and thus allowing for transcription. The Lac operon in bacteria is an example of a negatively-controlled operon (Figure 3).
A positive-repressible operon works in the opposite way. The operon is normally transcribed until a repressor/corepressor binds to the operator preventing transcription. The trp operon involved in the production of tryptophan is an example of a positively-controlled operon.
Common Bacterial Promoters
Researchers often incorporate the following promoters into plasmids to drive constitutive or inducible expression.
| Promoter | Expression | Description |
|---|---|---|
| T7 | Constitutive | Promoter from T7 bacteriophage; requires T7 RNA polymerase |
| Sp6 | Constitutive | Promoter from Sp6 bacteriophage; requires Sp6 RNA polymerase |
| lac | Constitutive | Promoter from Lac operon; constitutive in the absense of lac repressor (lacI or lacIq). Can be induced by IPTG or lactose |
| araBAD | Inducible by arabinose | Promoter of the arabinose metabolic operon |
| trp | Repressible by tryptophan | Promoter from E. coli tryptophan operon |
| Ptac | Constitutive | Hybrid promoter of lac and trp; constitutive in the absense of lac repressor (lacI or lacIq). Can be induced by IPTG or lactose |
Eukaryotic Promoters
Eukaryotic promoters are much more complex and diverse than prokaryotic promoters and span a wider range of DNA sequences. It is not unusual to have several regulatory elements, such as enhancers, several kilobases away from the transcription start site (TSS). Eukaryotic promoters are so complex in structure that the DNA tends to fold back on itself, which helps to explain how many physically distant DNA sequences can affect transcription of a given gene.
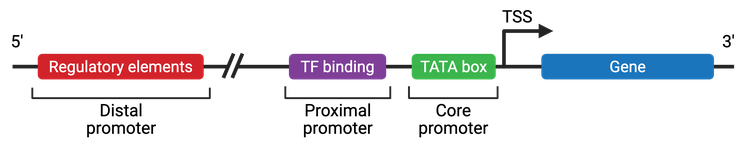
There are three main portions that make up a eukaryotic promoter: the core promoter, the proximal promoter, and the distal promoter (Figure 4).
Core Promoter
The core promoter region is located closest to the start codon and contains the RNA polymerase binding site, TATA box, and TSS. RNA polymerase will stably bind to this core promoter region and allow for the initiation of transcription of the template strand. The TATA box is a DNA sequence (TATAAA) within the core promoter region. The TATA-binding protein binds the TATA box and helps in the subsequent binding of the RNA polymerase. General transcription factors and histones (proteins that package DNA into nucleosomes) also bind the TATA box. Histone binding prevents the initiation of transcription, whereas transcription factors promote the initiation of transcription. The binding of RNA polymerase, several transcription factors, and other accessory proteins (such as activators and structural proteins) make up the transcription complex. The most 3' portion (closest to the gene's start codon) of the core promoter is the TSS, which is where transcription begins.
Proximal Promoter
Further upstream from the core promoter is the proximal promoter, which contains many primary regulatory elements. The proximal promoter is found approximately 250 base pairs upstream from the TSS and is the site where general transcription factors bind.
Distal Promoter
The final portion of the promoter region is called the distal promoter, which is upstream of the proximal promoter. The distal promoter also contains transcription factor binding sites, but mostly contains regulatory elements.
Common Eukaryotic Promoters
Researchers often incorporate the following promoters into plasmids to drive constitutive or inducible expression. Many other promoters can drive expression in specific cells or under different conditions.
| Promoter | Expression | Species | Description |
|---|---|---|---|
| Ac5 | Constitutive | Insect | Strong promoter from Drosophila actin 5c gene |
| Gal4/UAS | Specific | Insect | Requires UAS regulatory element and yeast Gal4 gene; often used in Drosophila |
| Polyhedrin | Constitutive | Insect | Strong promoter from baculovirus |
| CAG | Constitutive | Mammalian | Strong hybrid promoter; contains CMV early enhancer element and the chicken beta-actin promoter |
| CMV | Constitutive | Mammalian | Strong promoter from human cytomegalovirus |
| EF1a | Constituitve | Mammalian | Strong promoter from human elongation factor 1 alpha |
| PGK | Constitutive | Mammalian | Promoter from phospholycerate kinase gene |
| U6 | Constitutive | Mammalian | U6 nuclear promoter for small RNA expression (species-specific versions) |
| CaMV35S | Constitutive | Plant | Strong promoter from Cauliflower Mosaic Virus |
| Ubi | Constitutive | Plant | High-expression promoter from maize ubiquitin gene |
| GDS | Constitutive | Yeast | Very strong promoter from glyceraldehyde 3-phosphage dehydrogenase; also called TDH3 or GAPDH |
| TEF1 | Constitutive | Yeast | Yeast transcription elongation factor promoter |
| TRE | Inducible | Multiple | Tetracycline response element promoter; often optimized for mammalian systems |
Types of RNA Polymerases
Promoters control the binding of RNA polymerase to DNA to initiate the transcription of genes. There are three types of RNA polymerases that all transcribe different genes:
- RNA polymerase I — transcribes genes encoding ribosomal RNA (rRNA), which is a main component of a cell’s ribosome structure. Ribosomes are the site of protein syntehsis where mRNA is translated into a protein.
- RNA polymerase II — transcribes messenger RNA (mRNA), which is the RNA responsible for providing a stable template for the translation of a protein.
- RNA polymerase III — transcribes genes encoding transfer RNAs (tRNA), the adaptor molecules that are responsible for bringing amino acids to the ribosome when proteins are being synthesized. RNA Polymerase III also transcribes small RNAs, such as shRNAs and gRNAs.
Additional Resources
Content last reviewed: 22 October 2025


