We narrowed to 138,953 results for: LAS
-
Plasmid#20463DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only
-
pWZL Neo Myr Flag IHPK2
Plasmid#20538DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag SPHK2
Plasmid#20571DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag FRK
Plasmid#20488DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag CDK4
Plasmid#20448DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag MAP3K6
Plasmid#20519DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag RPS6KA6
Plasmid#20623DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pDONR223-VRK3
Plasmid#23687DepositorInsertVRK3 (VRK3 Human)
UseGateway donor vectorAvailable SinceJuly 30, 2010AvailabilityAcademic Institutions and Nonprofits only -
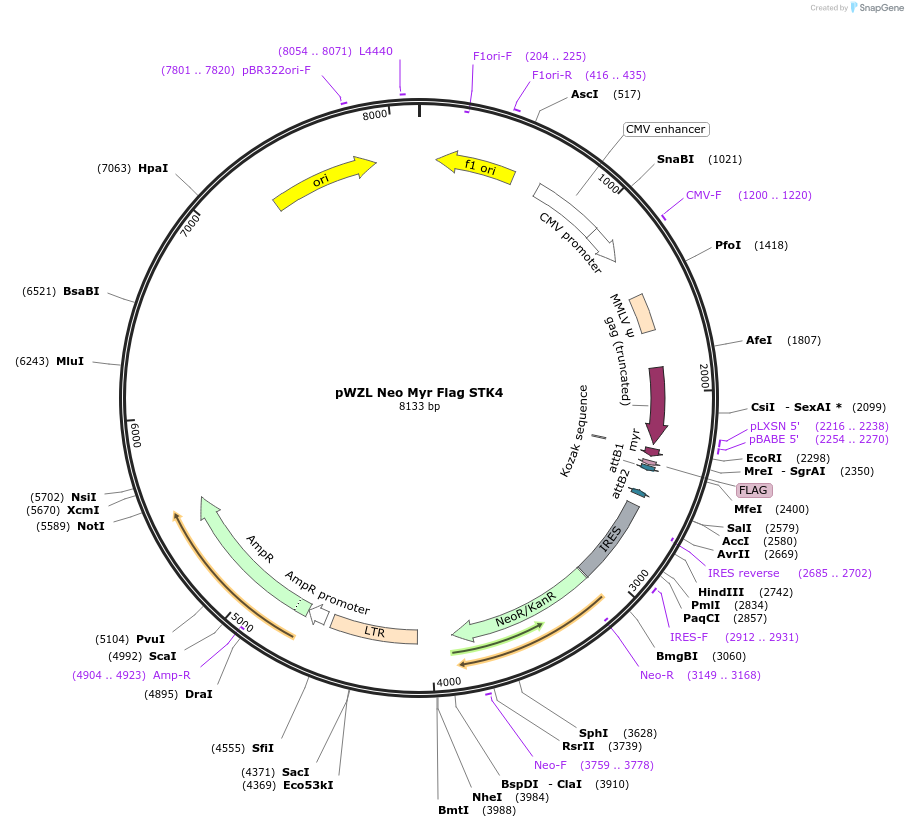
pWZL Neo Myr Flag STK4
Plasmid#20563DepositorInsertSTK4 (STK4 Human)
UseRetroviralTagsFlag and MyrExpressionMammalianMutationonly contains aa1-32 of STK4Available SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag ADCK5
Plasmid#20416DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag PCTK1
Plasmid#20560DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag PIK3R3
Plasmid#20575DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag PDXK
Plasmid#20566DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag CLK3
Plasmid#20465DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag MPP1
Plasmid#20542DepositorAvailable SinceMarch 24, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag CKMT2
Plasmid#20554DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pDONR223-RBKS
Plasmid#23681DepositorInsertRBKS (RBKS Human)
UseGateway donor vectorAvailable SinceJuly 30, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag ITPK1
Plasmid#20508DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag CSNK1G2
Plasmid#20471DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
pWZL Neo Myr Flag MOBKL1A
Plasmid#20503DepositorAvailable SinceOct. 27, 2009AvailabilityAcademic Institutions and Nonprofits only