We narrowed to 2,526 results for: Alb
-
Plasmid#24830DepositorAvailable SinceApril 14, 2011AvailabilityAcademic Institutions and Nonprofits only
-
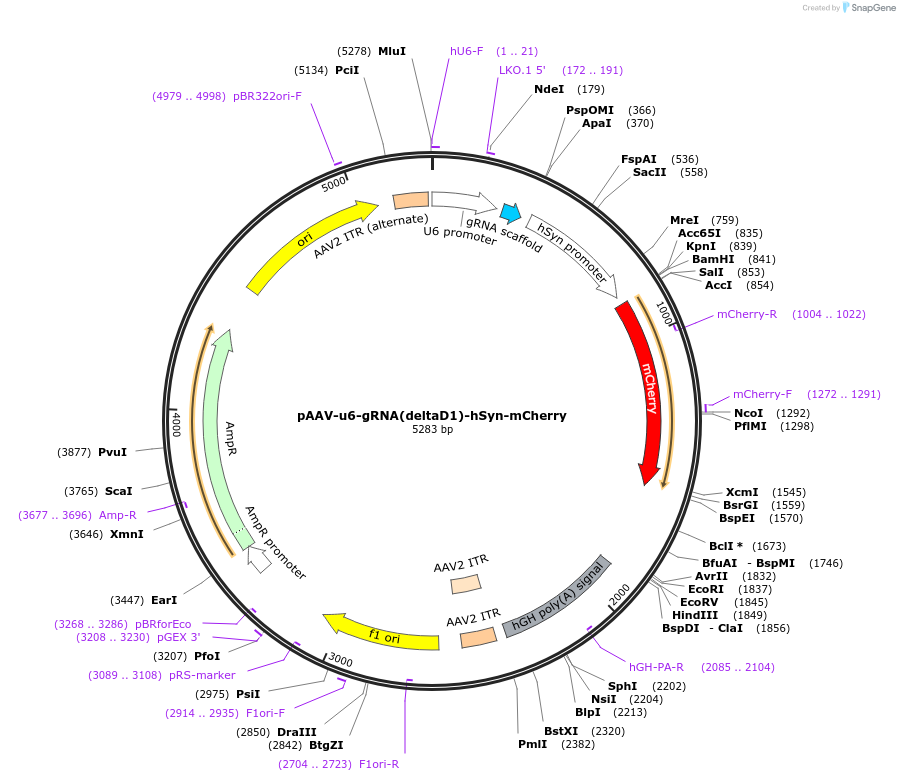
pAAV-u6-gRNA(deltaD1)-hSyn-mCherry
Plasmid#231400PurposeKnockdown of DRD1 across rodent speciesDepositorInsertsgRNA(DRD1.2)
UseAAV and CRISPRExpressionMammalianPromoteru6Available SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
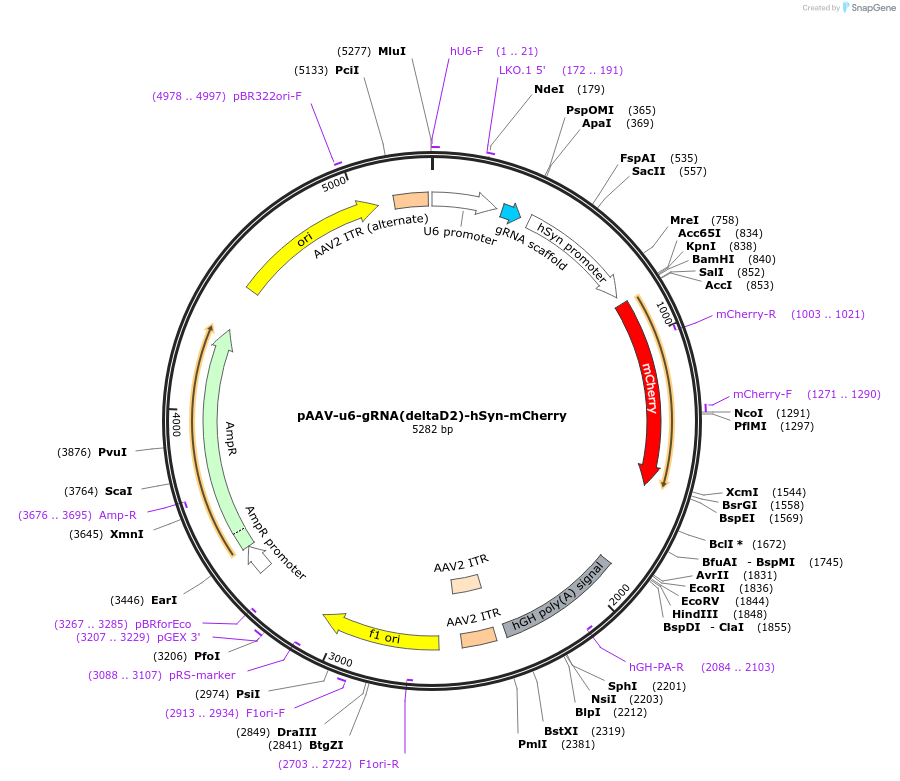
pAAV-u6-gRNA(deltaD2)-hSyn-mCherry
Plasmid#231401PurposeKnockdown of DRD2 across rodent speciesDepositorInsertsgRNA(DRD2.1)
UseAAV and CRISPRExpressionMammalianPromoterU6Available SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
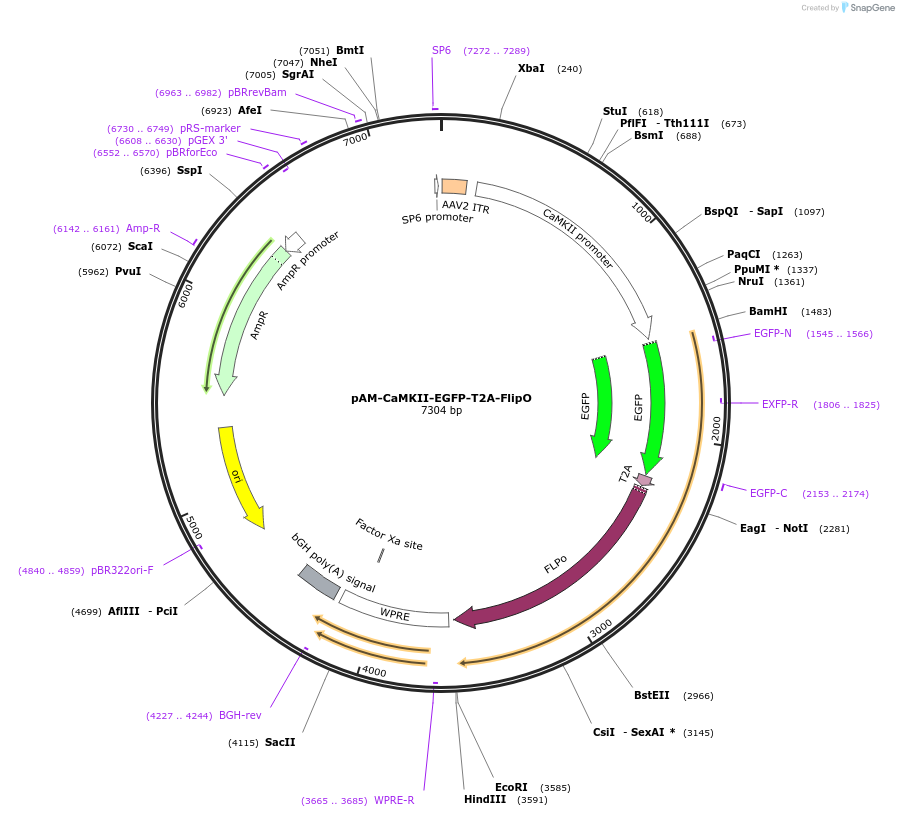
pAM-CaMKII-EGFP-T2A-FlipO
Plasmid#232791PurposeAAV2 vector containing EGFP and FlipO sequences separated by T2A peptide under the control of CaMKIIA promoter.DepositorInsertCaMKII-EGFP-T2A-FlipO
UseAAVAvailable SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
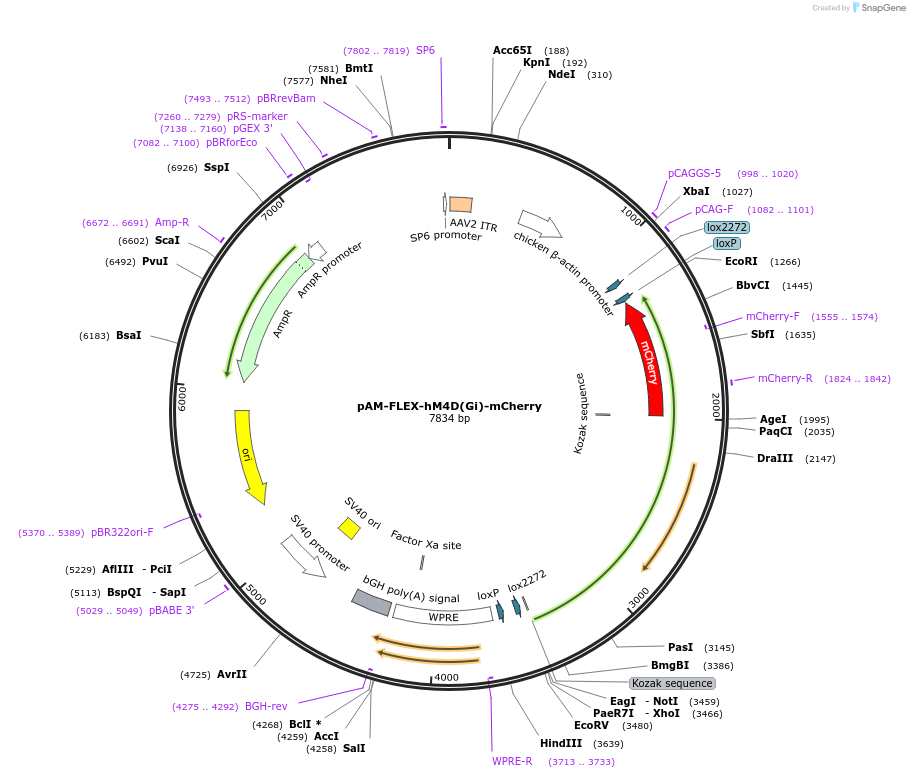
pAM-FLEX-hM4D(Gi)-mCherry
Plasmid#232782PurposeAAV2 vector containing Gi-coupled hM4D DREADD fused with mCherry inside a double floxed (FLEX) cassette under the control of CBA promoter.DepositorInserthM4D(Gi)-mCherry
UseAAVAvailable SinceApril 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
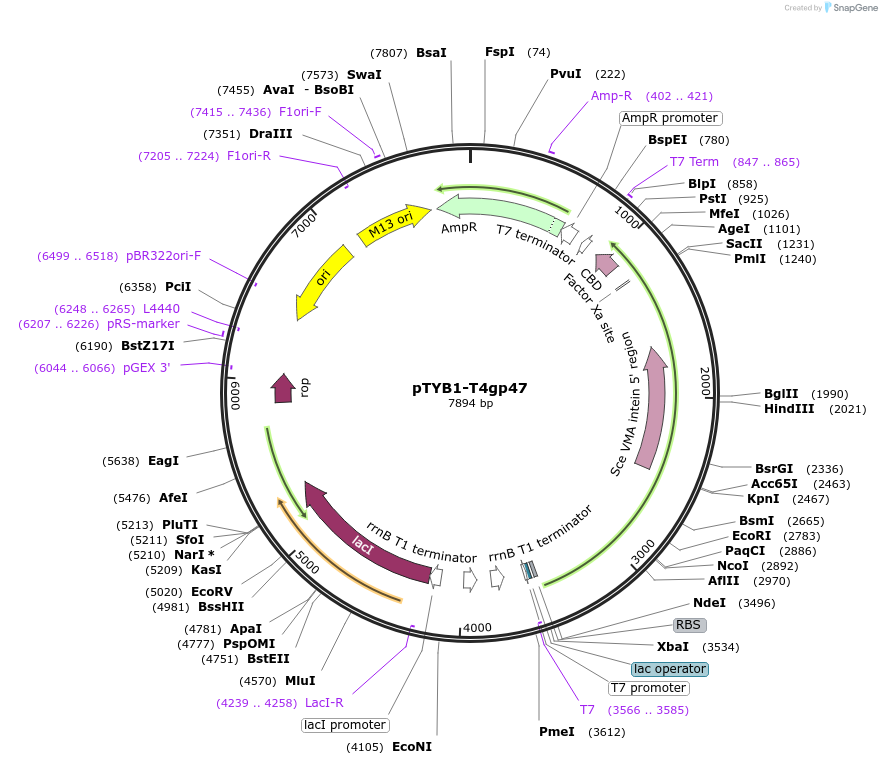
pTYB1-T4gp47
Plasmid#228212PurposeExpression in BL21(DE3) of T4 SbcD-like subunit of palindrome specific endonucleaseDepositorInsert47 (47 Escherichia phage T4)
TagsSce VMA intein-chitin binding domainExpressionBacterialPromoterT7Available SinceDec. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
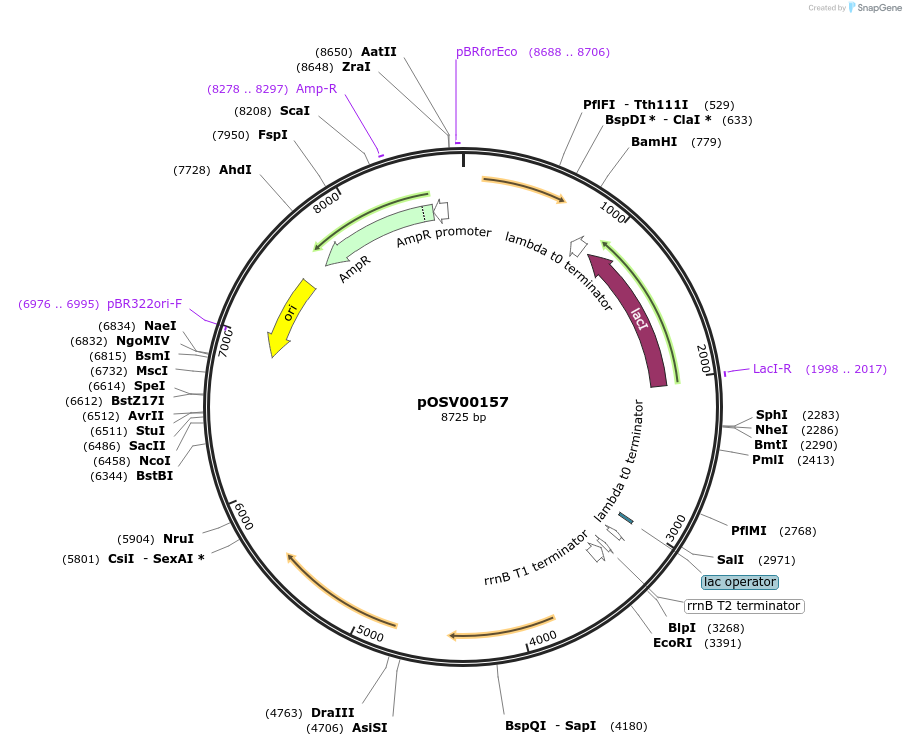
pOSV00157
Plasmid#222937PurposeKill switch plasmid for DNA sensor construction using Bacillus subtilisDepositorInsertTxpA-RatA toxin-antitoxin
ExpressionBacterialPromoterPhyperspankAvailable SinceDec. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
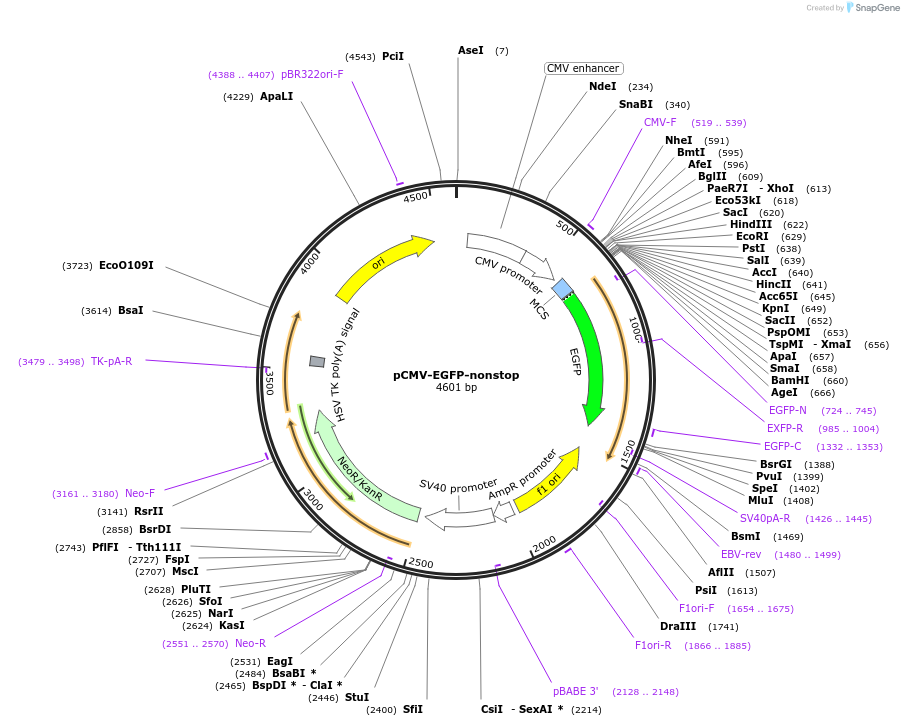
pCMV-EGFP-nonstop
Plasmid#226406PurposeExpresses EGFP without a stop codon that produces a GFP-tagged readthrough RQC substrate.DepositorInsertEGFP without stop codon
ExpressionMammalianPromoterCMVAvailable SinceNov. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
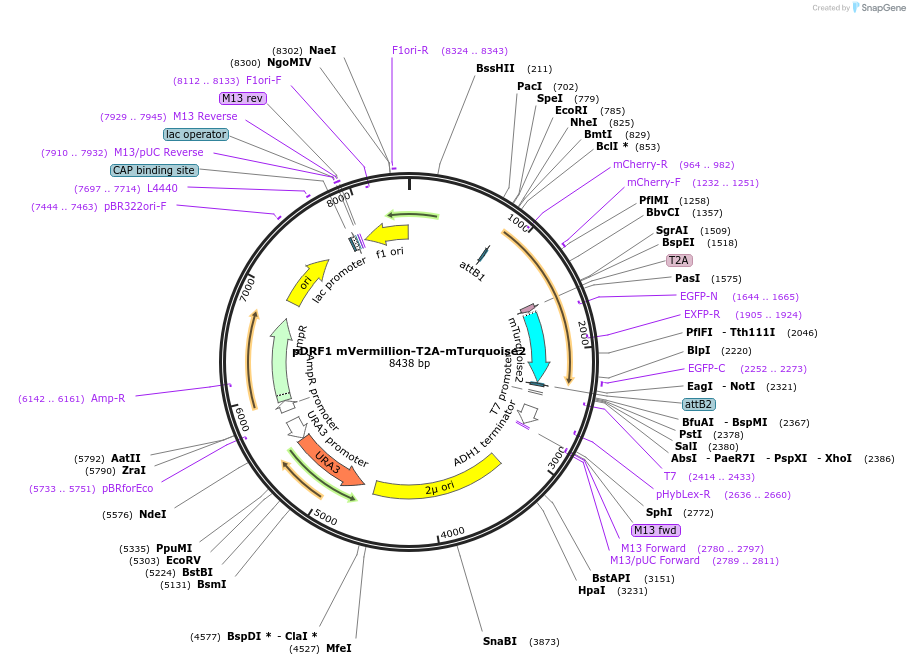
pDRF1 mVermillion-T2A-mTurquoise2
Plasmid#219847Purposeequimolar expression of mVermillion and mTurquoise2 in budding yeastDepositorInsertmVermillion-T2A-mTurquoise2
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
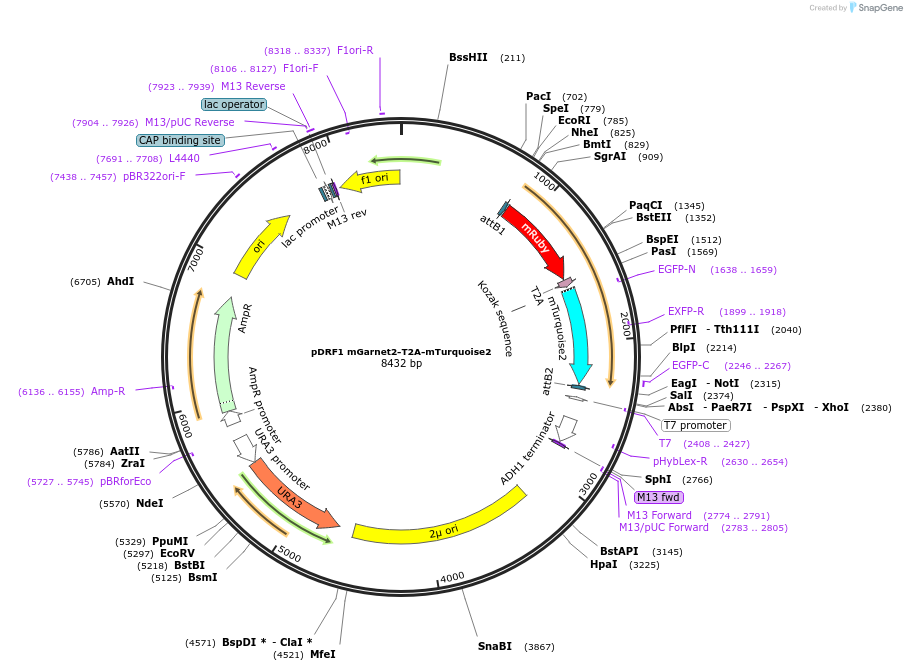
pDRF1 mGarnet2-T2A-mTurquoise2
Plasmid#219850Purposeequimolar expression of mGarnet2 and mTurquoise2 in budding yeastDepositorInsertmGarnet2-T2A-mTurquoise2
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
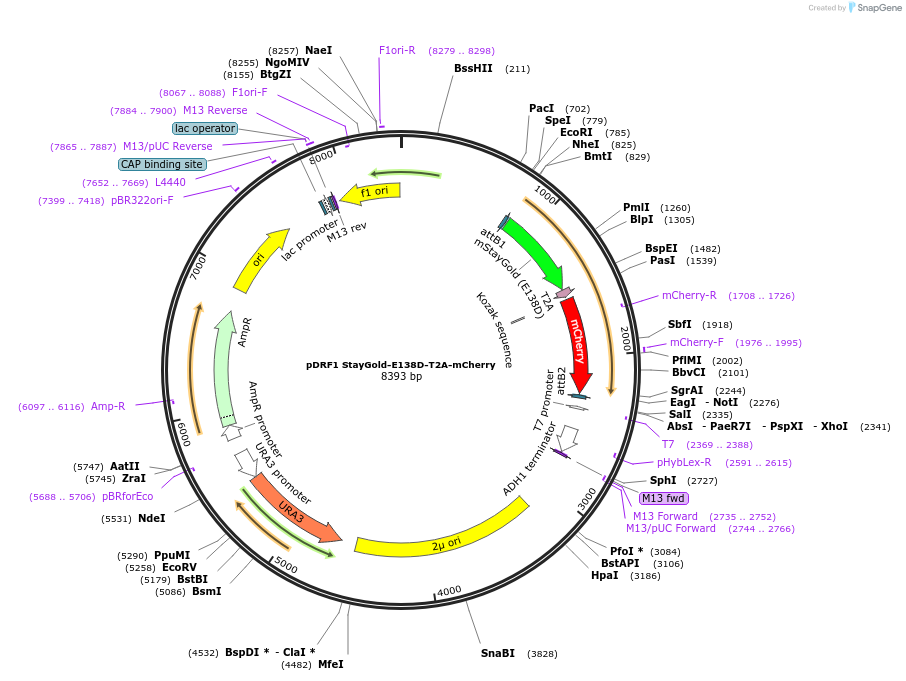
pDRF1 StayGold-E138D-T2A-mCherry
Plasmid#219833Purposeequimolar expression of Staygold-E138D and mCherry in budding yeastDepositorInsertStayGold-E138D-T2A-mCherry
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
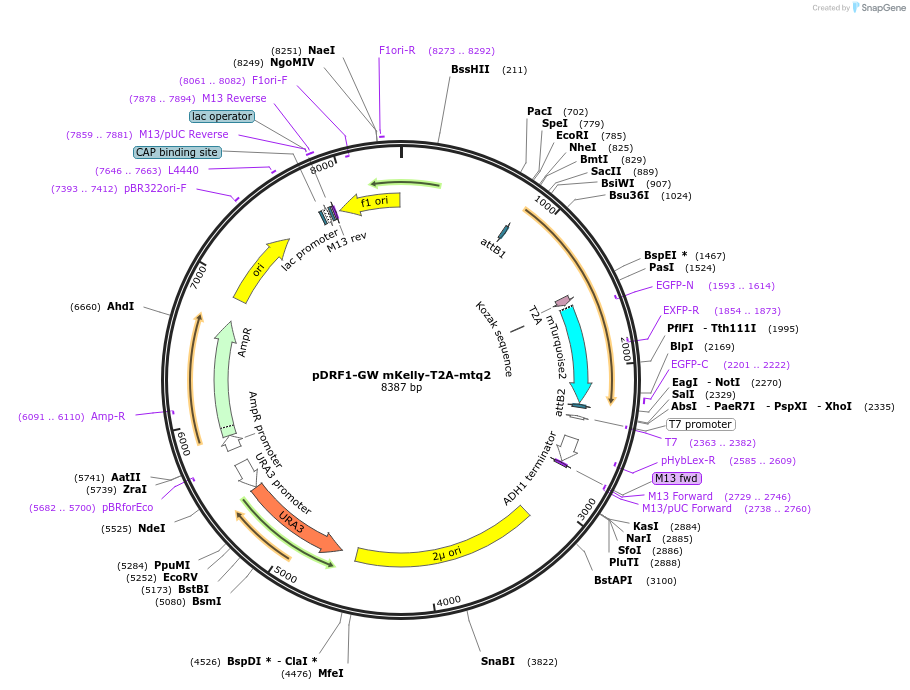
pDRF1-GW mKelly-T2A-mtq2
Plasmid#205506PurposeExpression of mKelly-T2A-mtq2 in yeastDepositorInsertmKelly-T2A-mtq2
ExpressionYeastAvailable SinceJuly 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
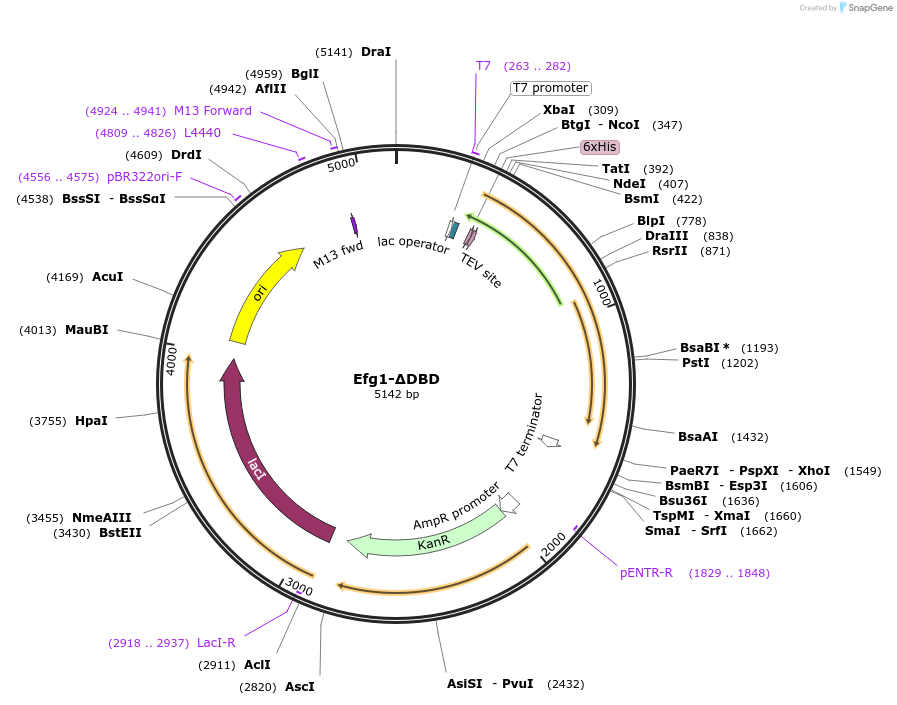
Efg1-ΔDBD
Plasmid#216396PurposeExpresses 6xHis-tagged N- and C-terminal prion-like domains of Efg1DepositorInsertEfg1-ΔDBD
Tags6xHis-tagsExpressionBacterialMutationdeleted amino acids 182-356Available SinceApril 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
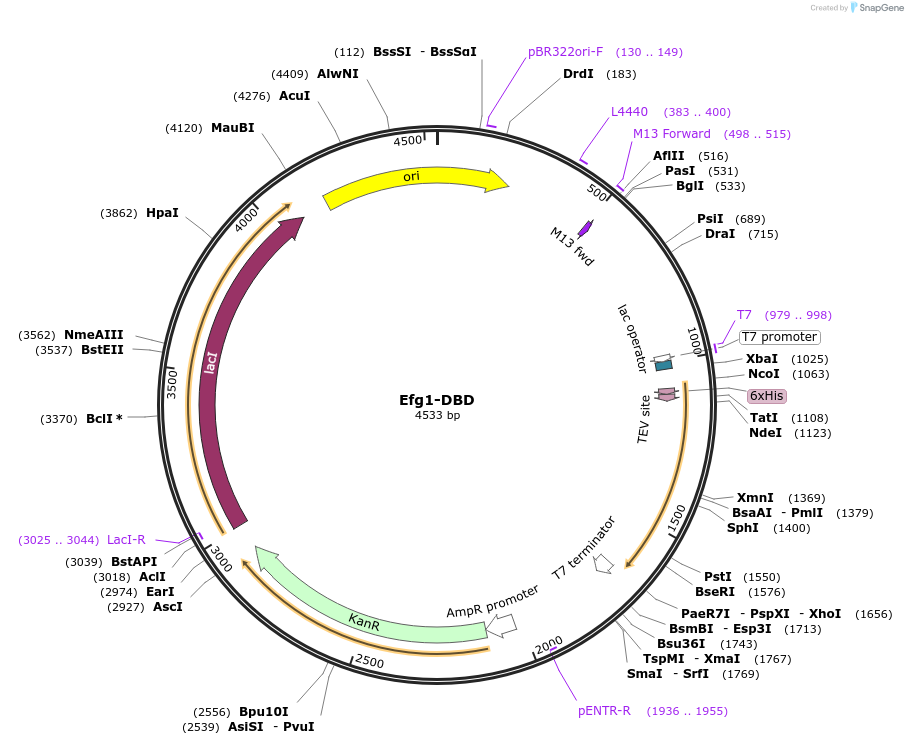
Efg1-DBD
Plasmid#216399PurposeExpresses 6xHis-tagged DBD of Efg1DepositorInsertEfg1-DBD
Tags6xHis-tagsExpressionBacterialMutationdeleted amino acids 2-181 and 357-554Available SinceApril 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
pDRF1-GW miFP663-T2A-mtq2
Plasmid#205504PurposeExpression of miFP663-T2A-mtq2 in yeastDepositorInsertmiFP663-T2A-mtq2
ExpressionYeastAvailable SinceSept. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
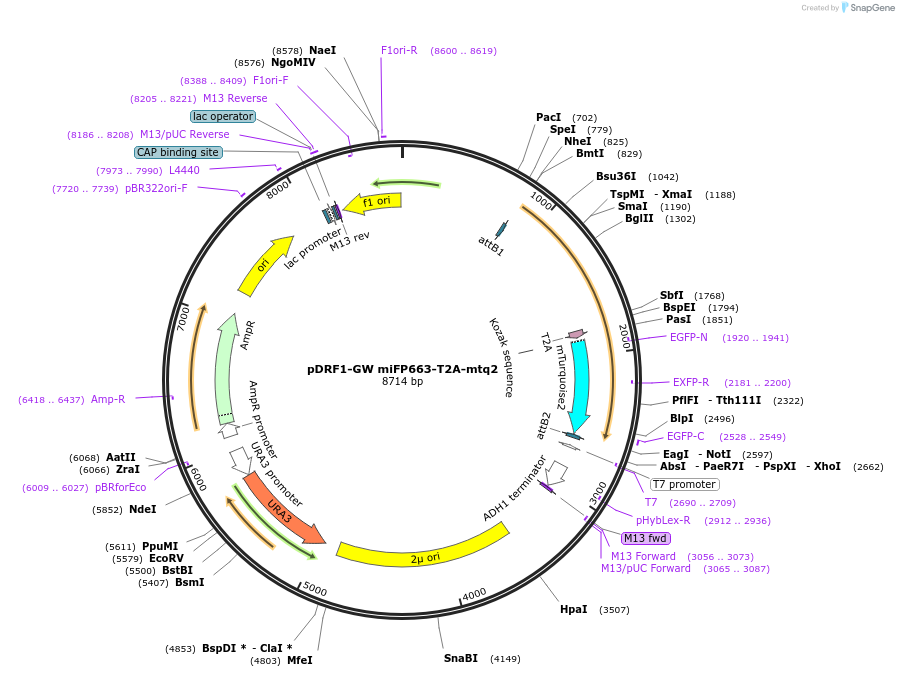
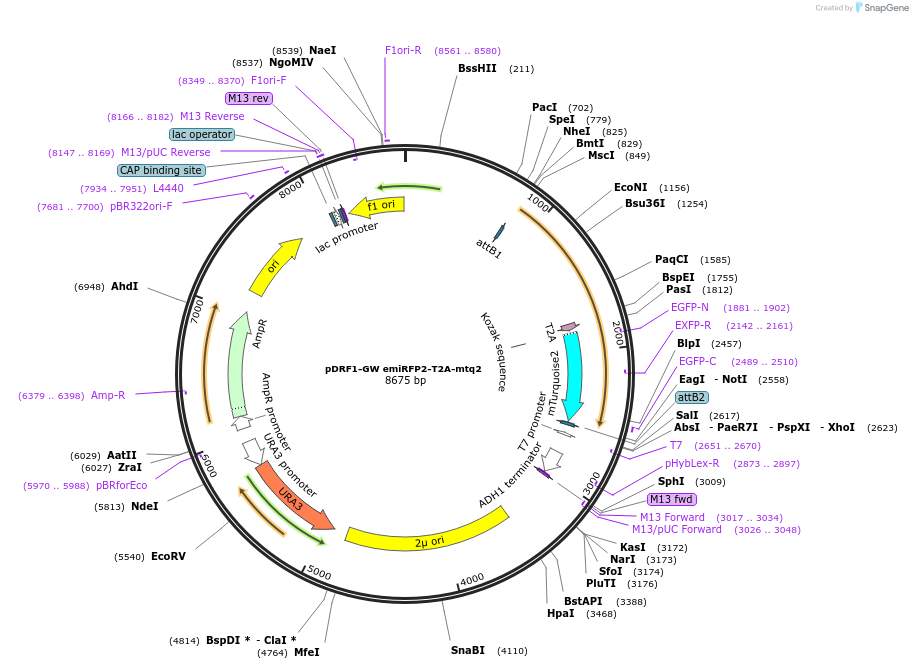
pDRF1-GW emiRFP2-T2A-mtq2
Plasmid#205497PurposeExpression of emiRFP2-T2A-mtq2 in yeastDepositorInsertemiRFP2-T2A-mtq2
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
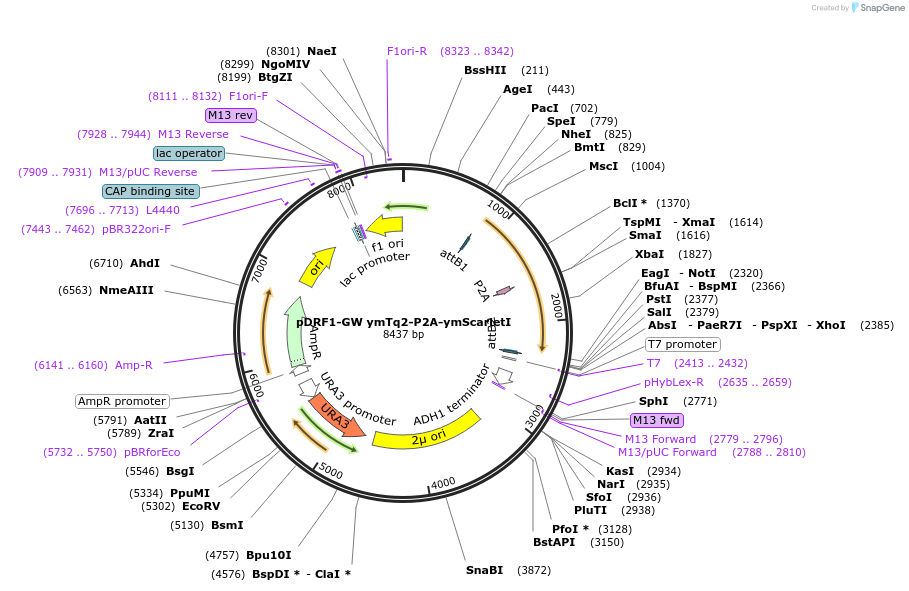
pDRF1-GW ymTq2-P2A-ymScarletI
Plasmid#205500PurposeExpression of ymTq2-P2A-ymScarletI in yeastDepositorInsertymTq2-P2A-ymScarletI
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
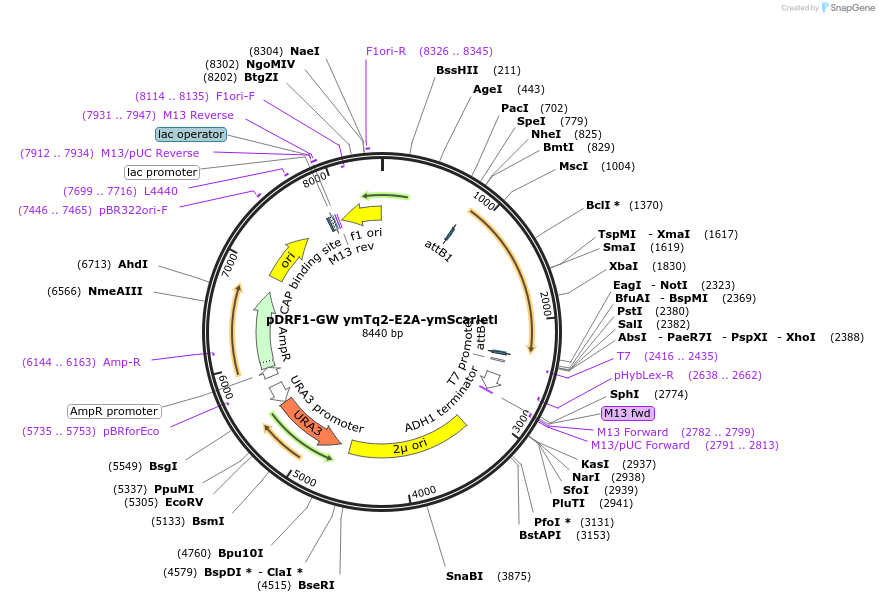
pDRF1-GW ymTq2-E2A-ymScarletl
Plasmid#205510PurposeExpression of ymTq2-E2A-ymScarletl in yeastDepositorInsertymTq2-E2A-ymScarletl
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
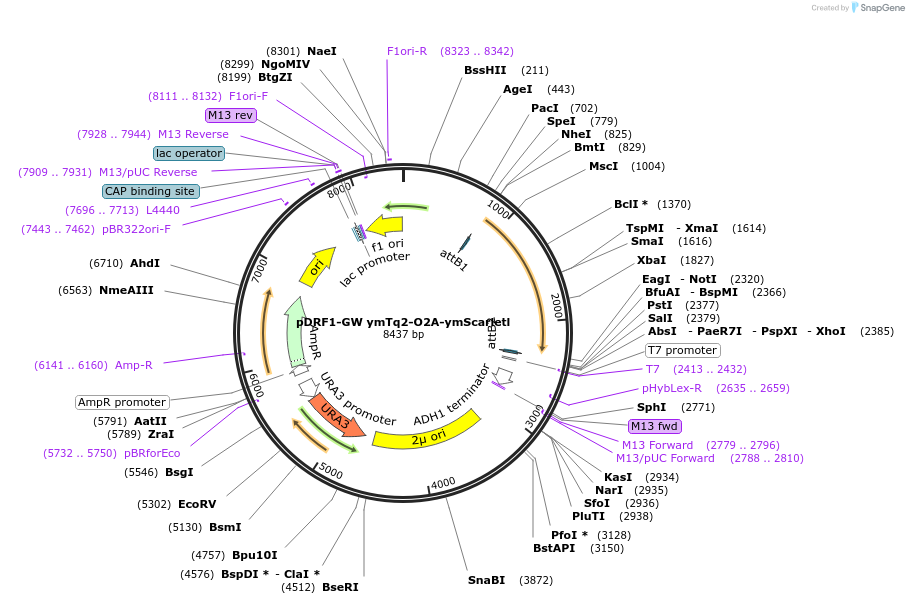
pDRF1-GW ymTq2-O2A-ymScarletl
Plasmid#205509PurposeExpression of ymTq2-O2A-ymScarletl in yeastDepositorInsertymTq2-O2A-ymScarletl
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
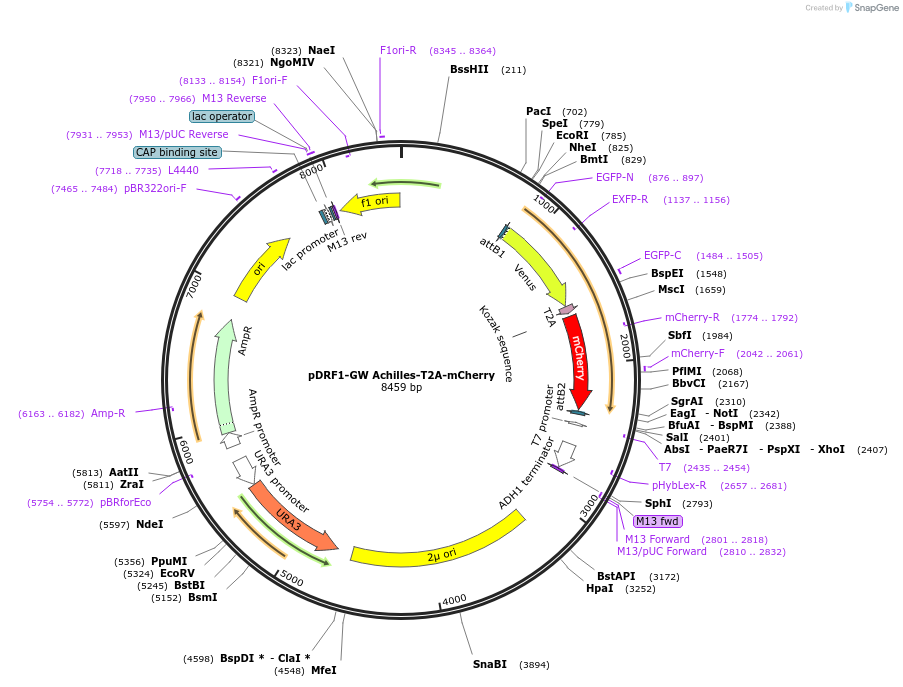
pDRF1-GW Achilles-T2A-mCherry
Plasmid#205499PurposeExpression of Achilles-T2A-mCherry in yeastDepositorInsertAchilles-T2A-mCherry
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
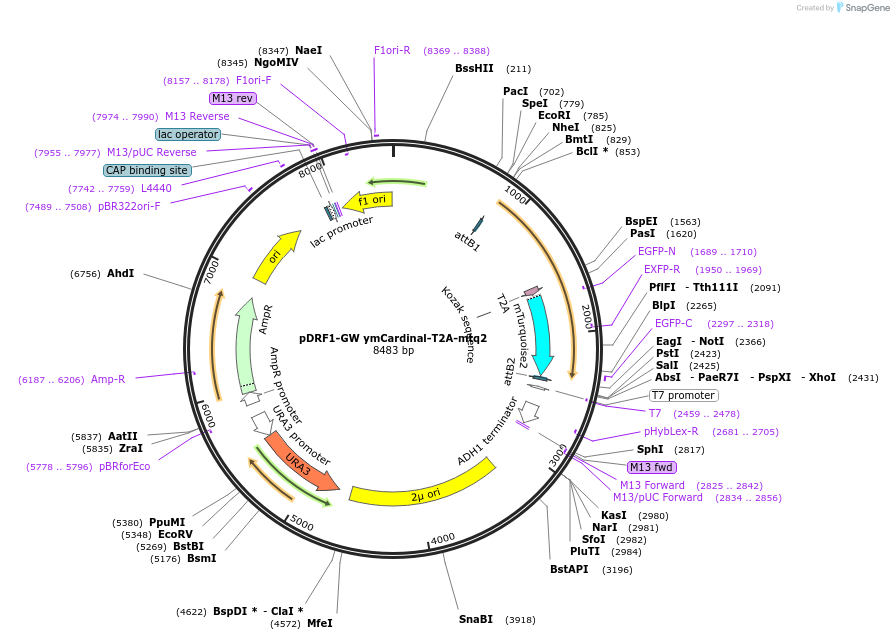
pDRF1-GW ymCardinal-T2A-mtq2
Plasmid#205508PurposeExpression of ymCardinal-T2A-mtq2 in yeastDepositorInsertymCardinal-T2A-mtq2
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
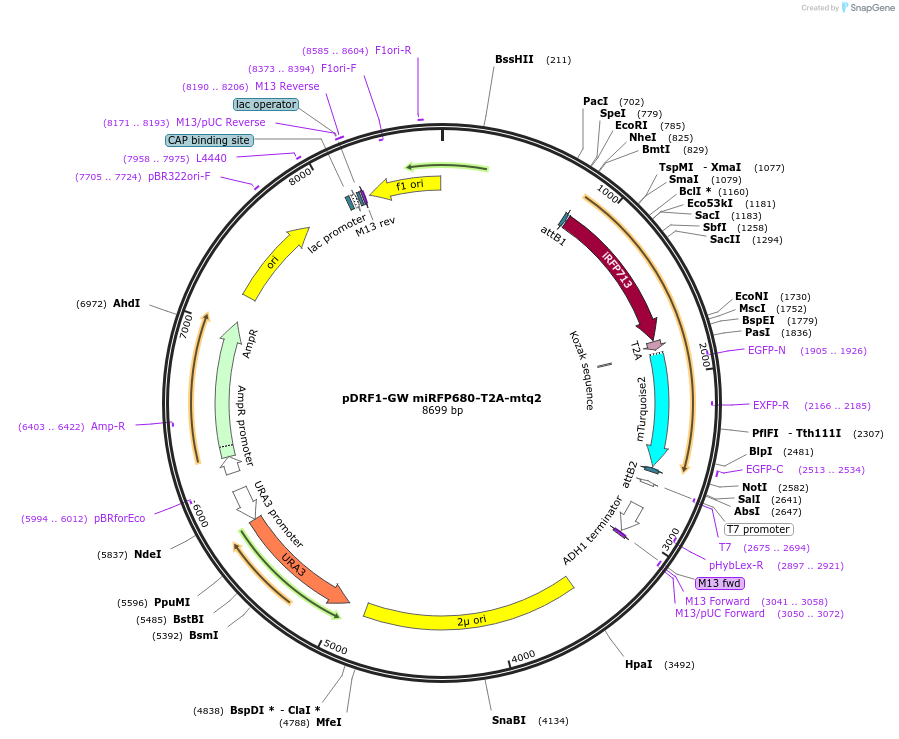
pDRF1-GW miRFP680-T2A-mtq2
Plasmid#205502PurposeExpression of miRFP680-T2A-mtq2 in yeastDepositorInsertmiRFP680-T2A-mtq2
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
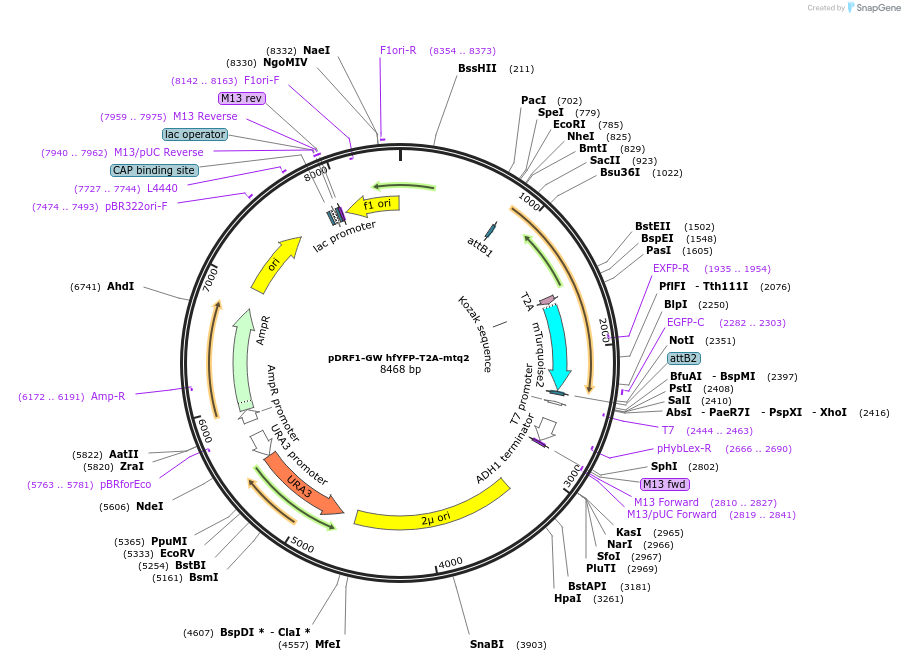
pDRF1-GW hfYFP-T2A-mtq2
Plasmid#205496PurposeExpression of hfYFP-T2A-mtq2 in yeastDepositorInserthfYFP-T2A-mtq2
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
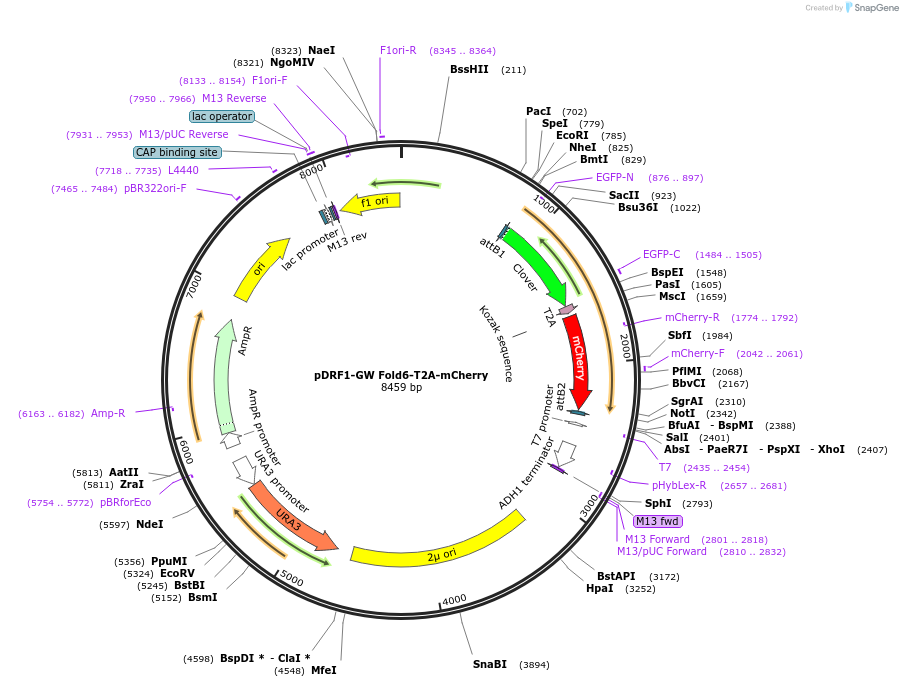
pDRF1-GW Fold6-T2A-mCherry
Plasmid#205492PurposeExpression of Fold6-T2A-mCherry in yeastDepositorInsertFold6-T2A-mCherry
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
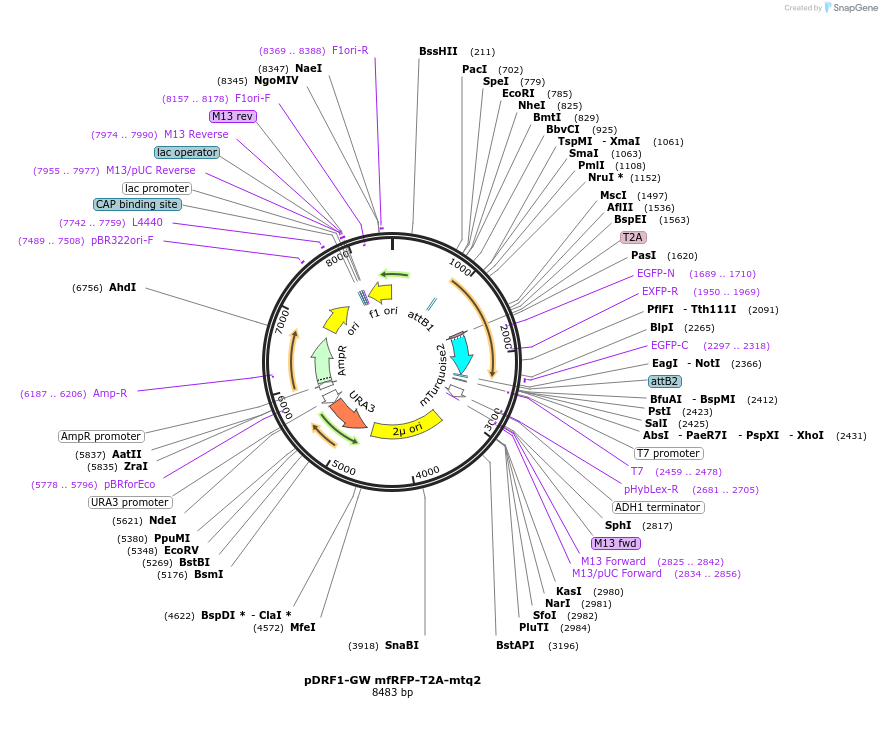
pDRF1-GW mfRFP-T2A-mtq2
Plasmid#205494PurposeExpression of mfRFP-T2A-mtq2 in yeastDepositorInsertmfRFP-T2A-mtq2
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
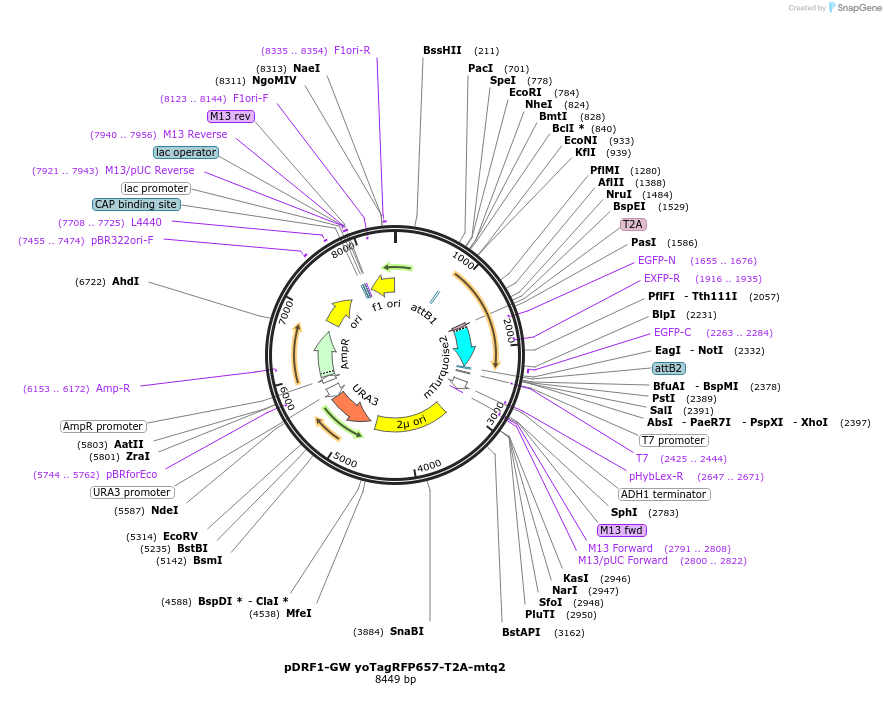
pDRF1-GW yoTagRFP657-T2A-mtq2
Plasmid#205505PurposeExpression of yoTagRFP657-T2A-mtq2 in yeastDepositorInsertyoTagRFP657-T2A-mtq2
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
pDRF1-GW miRFP2-T2A-mtq2
Plasmid#205503PurposeExpression of miRFP2-T2A-mtq2 in yeastDepositorInsertmiRFP2-T2A-mtq2
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
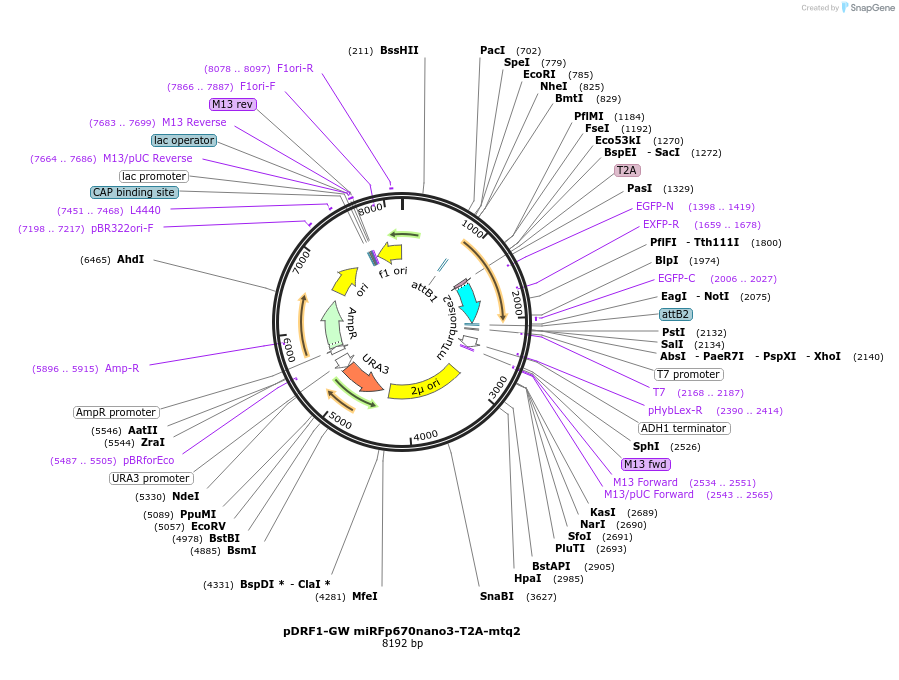
pDRF1-GW miRFp670nano3-T2A-mtq2
Plasmid#205501PurposeExpression of miRFp670nano3-T2A-mtq2 in yeastDepositorInsertmiRFp670nano3-T2A-mtq2
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
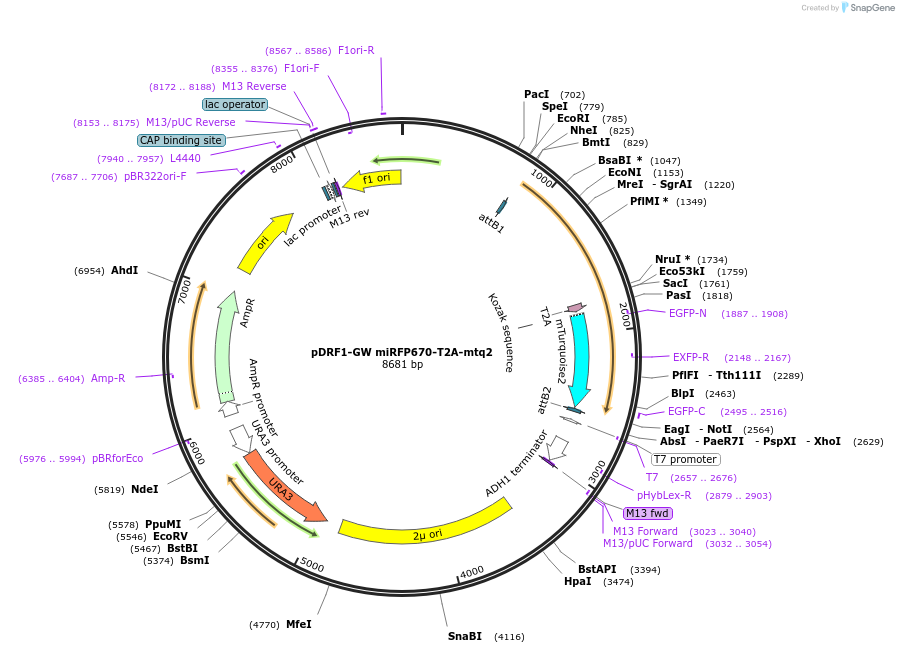
pDRF1-GW miRFP670-T2A-mtq2
Plasmid#205491PurposeExpression of miRFP670-T2A-mtq2 in yeastDepositorInsertmiRFP670-T2A-mtq2
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
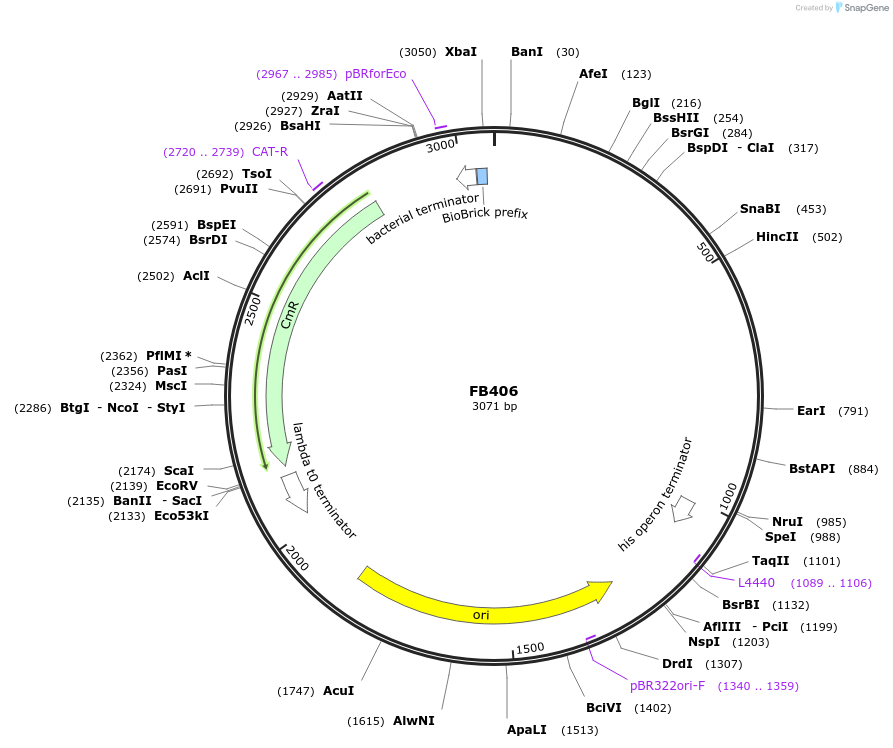
FB406
Plasmid#203627PurposePromoter of the TAKA-amylase A gene from A. oryzae, maltose/starch-inducible.DepositorInsertPamyB
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
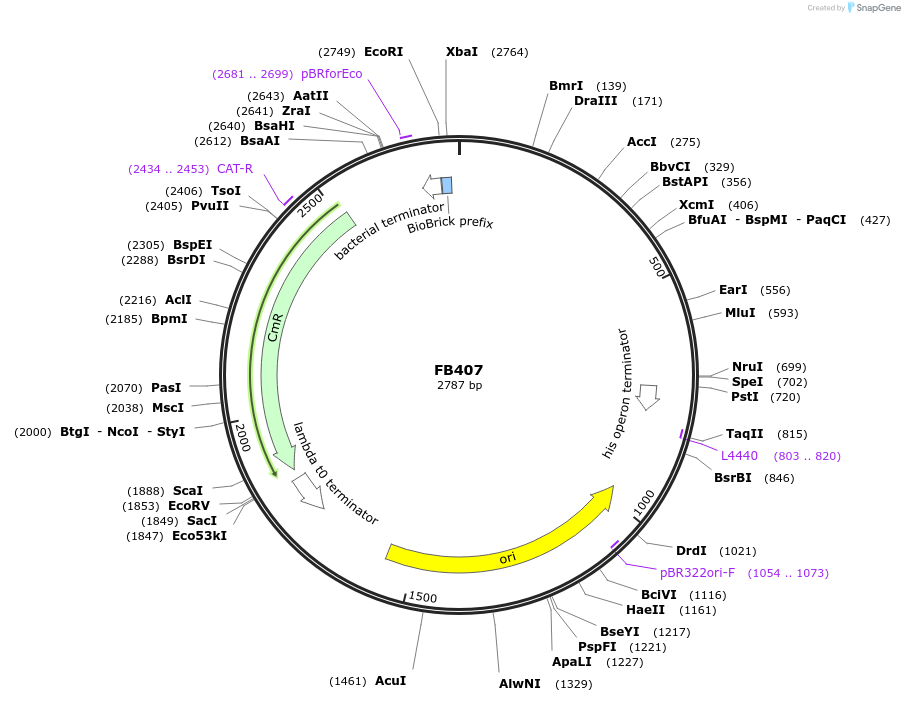
FB407
Plasmid#203628PurposePromoter of the elongation factor 1-alpha gene from P. digitatum (PDIG_59570).DepositorInsertPef1A
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
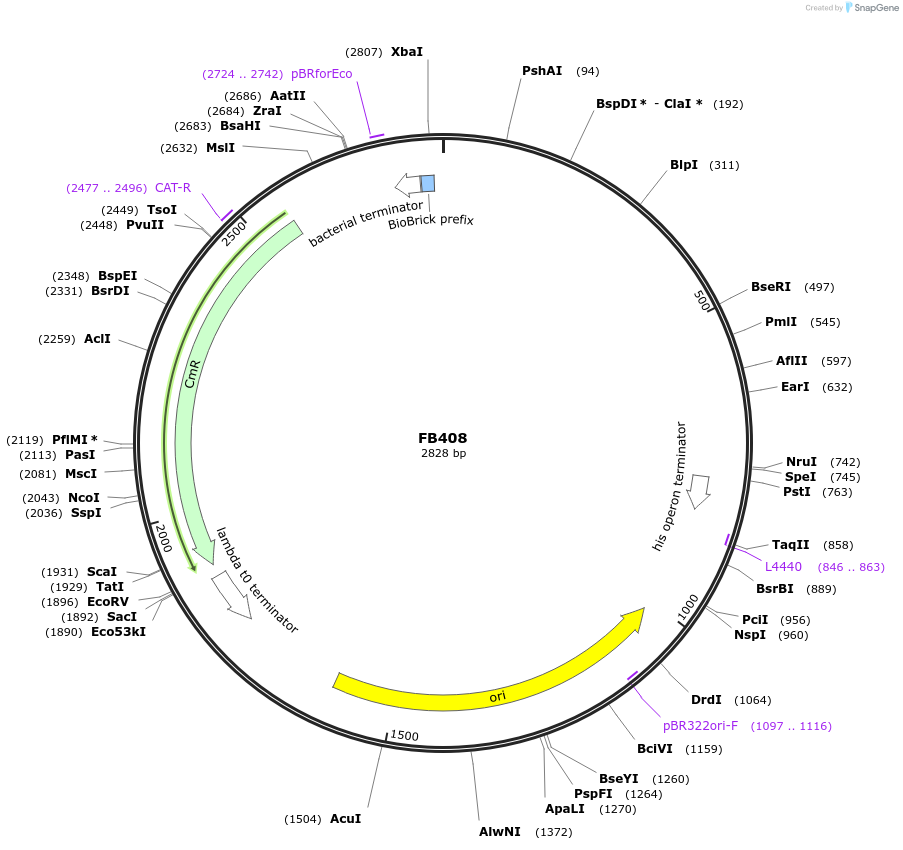
FB408
Plasmid#203629PurposePromoter of the ubiquitin ligase gene from P. digitatum (PDIG_07760).DepositorInsertP07760
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
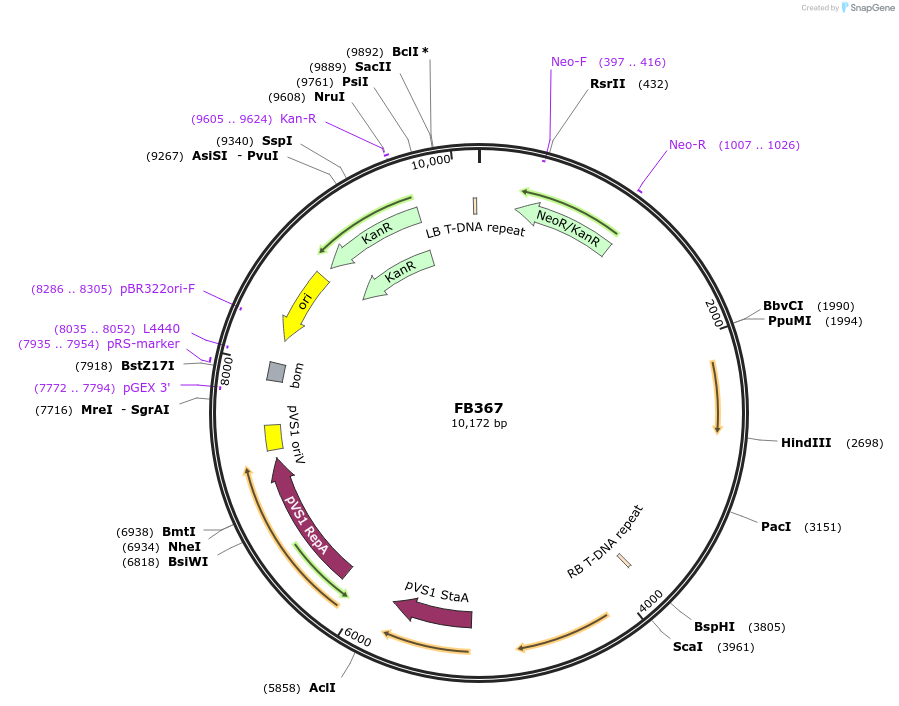
FB367
Plasmid#203630PurposeModule for the expression of geneticin resistance and Nanoluciferase.DepositorInsertPgpdA:Nluc:Ttub::PtrpC:nptII:Ttub
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
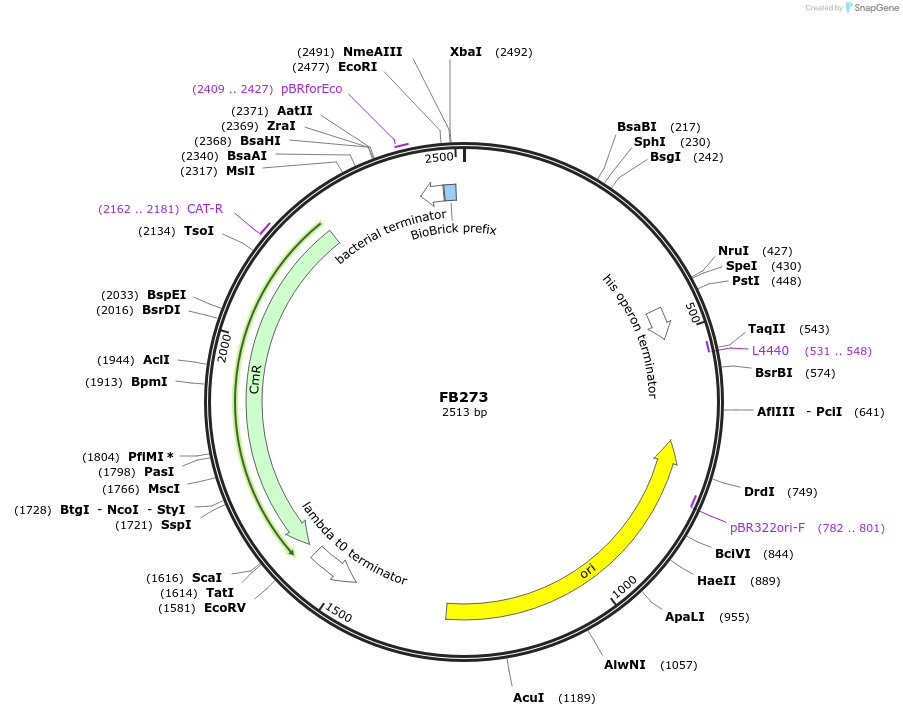
FB273
Plasmid#203612PurposeTerminator of pyr4 gene from T. reesei.DepositorInsertTpyr4
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
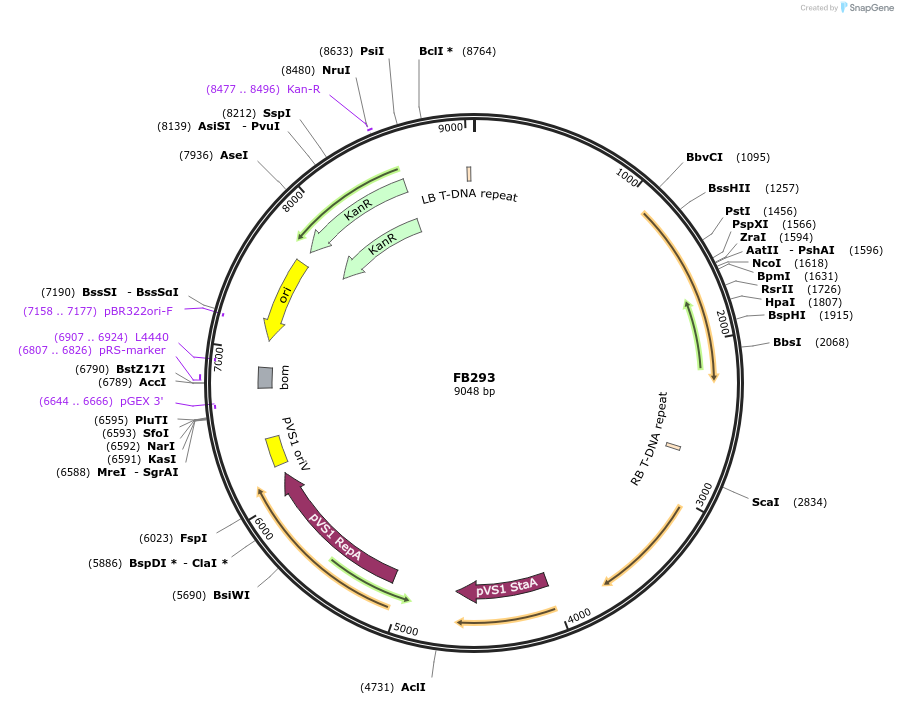
FB293
Plasmid#203613PurposeAssembly of the transcriptional unit for the auxotrophy marker pyr4 from T. reseei.DepositorInsertTU_pyr4
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
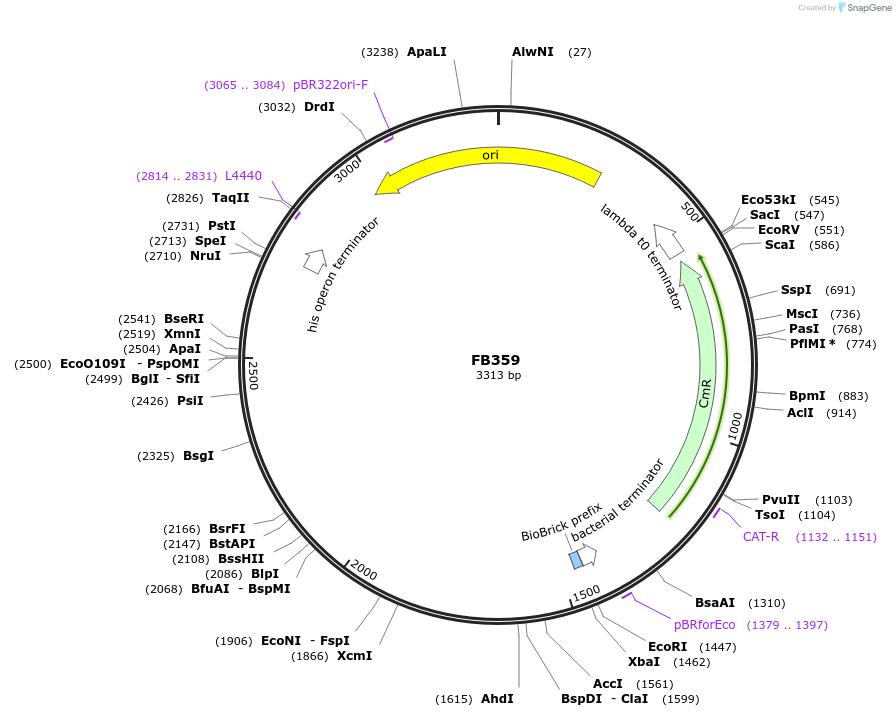
FB359
Plasmid#203614Purpose5' upstream region of pyrG gene in P. digitatum.DepositorInsert5' upstream Pdig pyrG
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
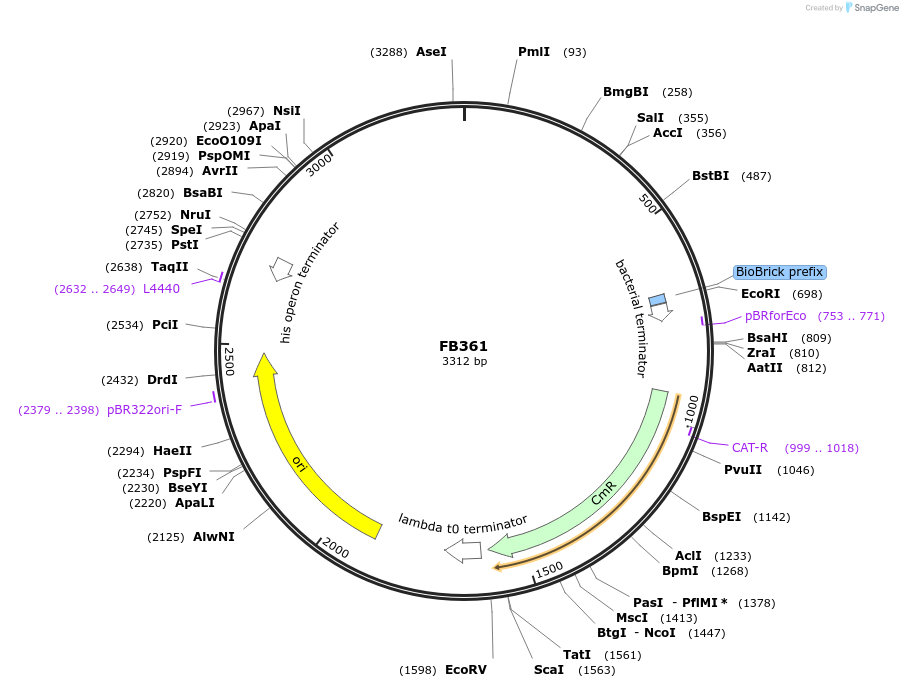
FB361
Plasmid#203615Purpose3' downstream region of pyrG gene in P. digitatum.DepositorInsert3' downstream Pdig pyrG
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
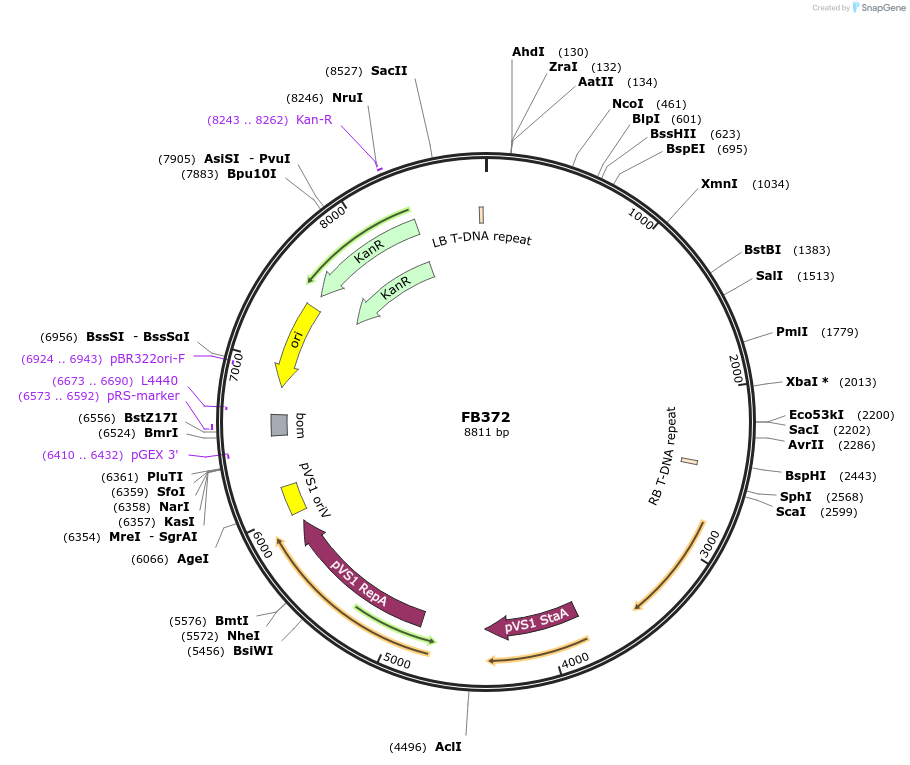
FB372
Plasmid#203616PurposeAssembly for pyrG deletion in P. digitatum.DepositorInsertFB359+FB361
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
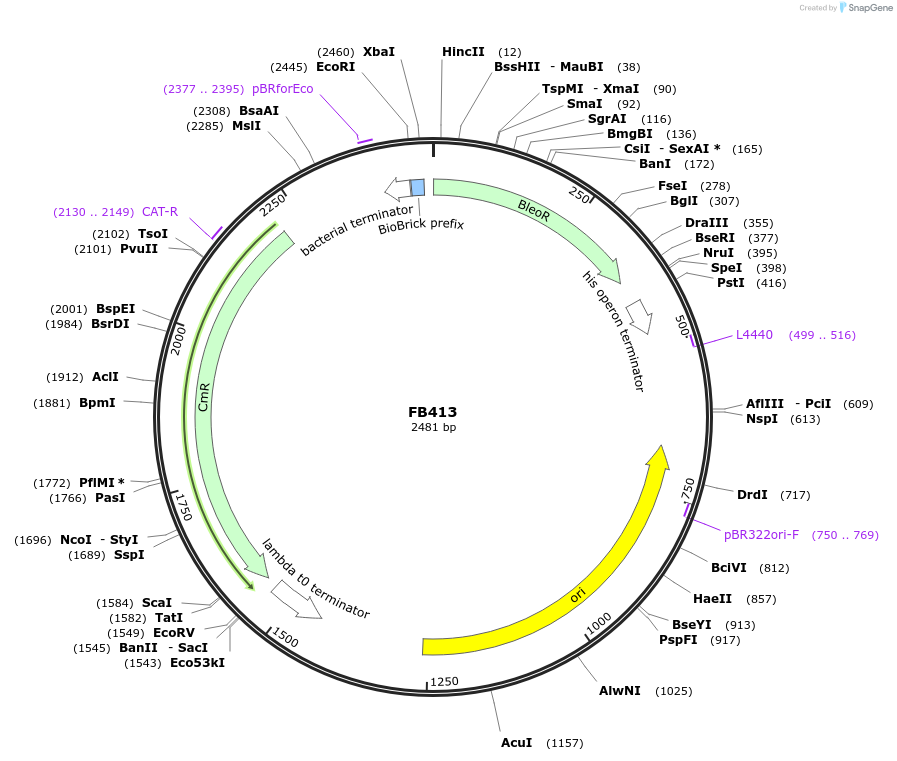
FB413
Plasmid#203617PurposeCoding sequence for phleomycin resistance.DepositorInsertble
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
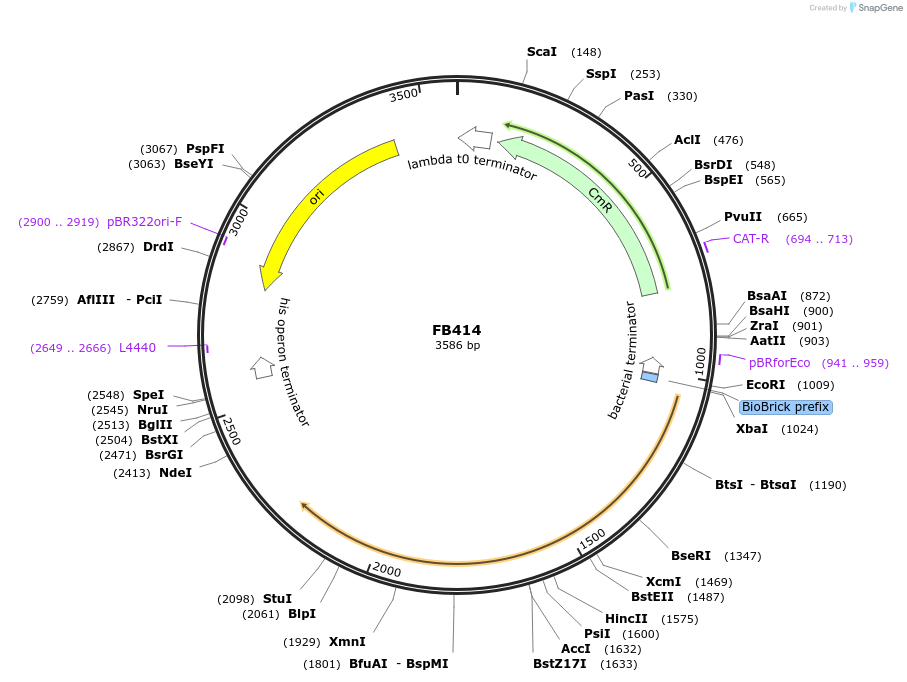
FB414
Plasmid#203618PurposeCoding sequence for terbinafine resistance.DepositorInsertergA
UseSynthetic BiologyAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only