We narrowed to 13,611 results for: Mpl;
-
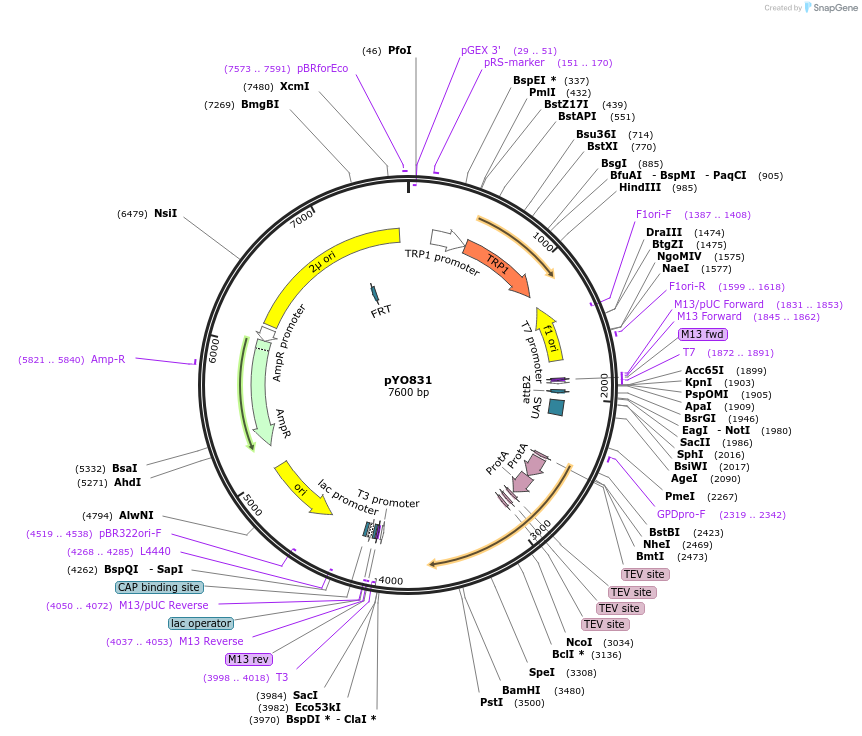
Plasmid#235754PurposeProtein expression of ZZ-ScVPS38 (1-212F)DepositorInsertVPS38
TagsZZ-3xTEVExpressionYeastMutationaa 1-212FAvailable SinceMay 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
pYO225
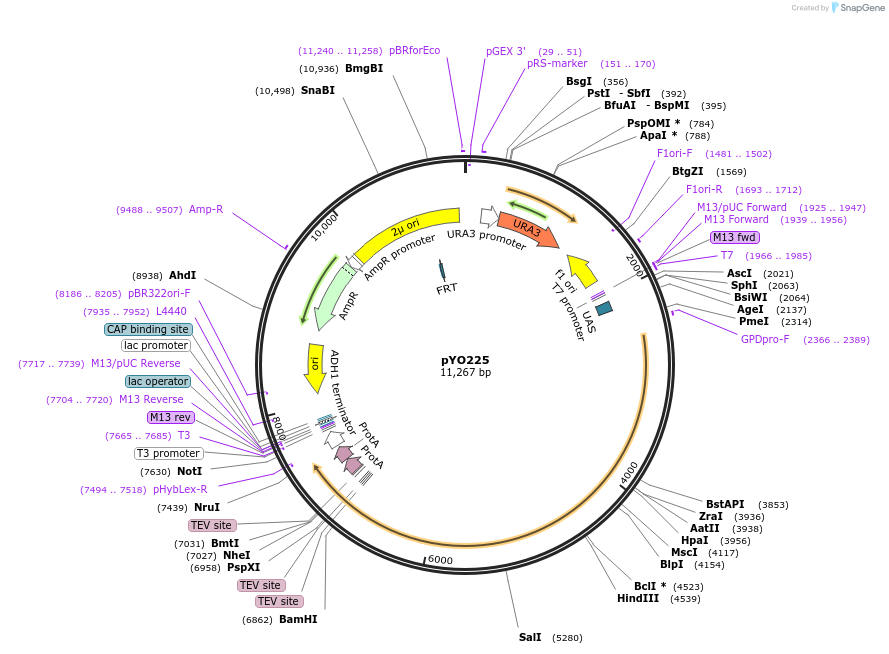
Plasmid#235729PurposeProtin expression of FL yeast VPS15 -ZZ in yeastDepositorInsertVPS15
Tags3xTEV-ZZExpressionYeastAvailable SinceMay 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
pYO290
Plasmid#235731PurposeProtein expression in FL yeast VPS34 D731N mutant in yeastDepositorInsertVPS34
ExpressionYeastMutationS2A, D731NAvailable SinceMay 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
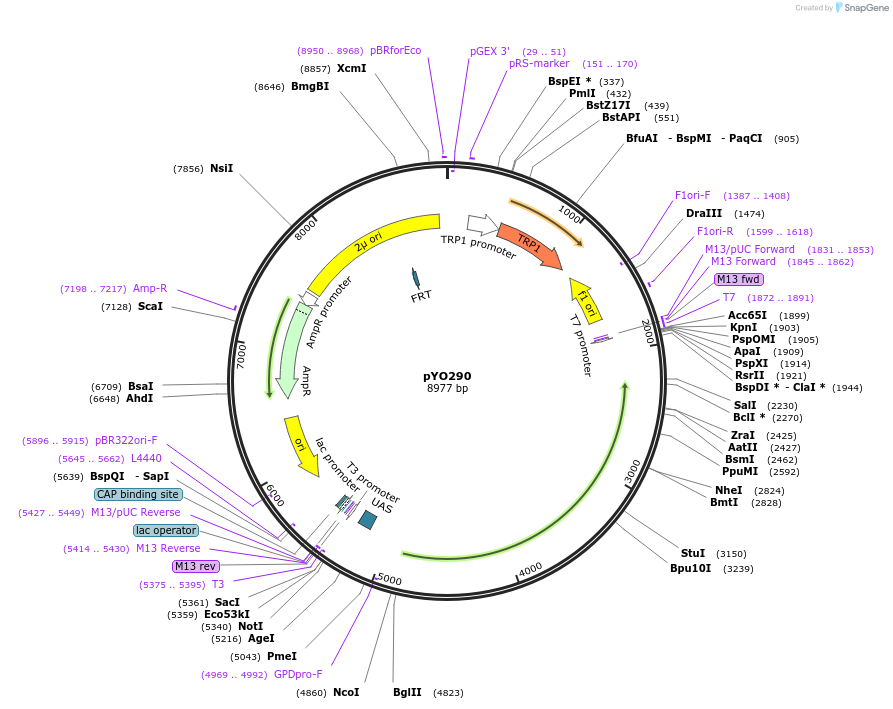
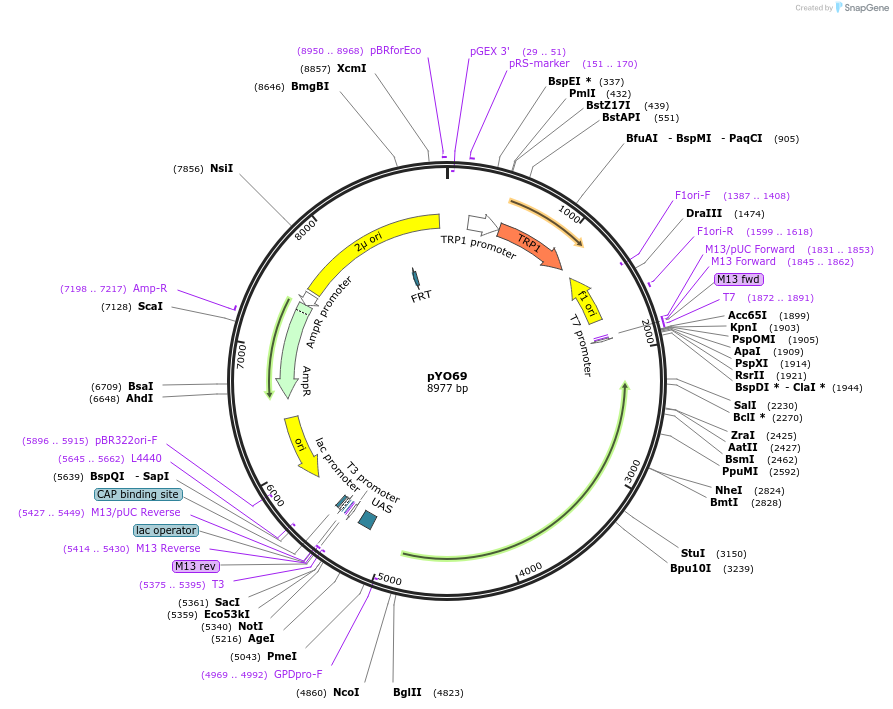
pYO69
Plasmid#235747PurposeProtein expression of FL ScVPS34 in yeastDepositorInsertVPS34
ExpressionYeastMutationS2AAvailable SinceMay 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
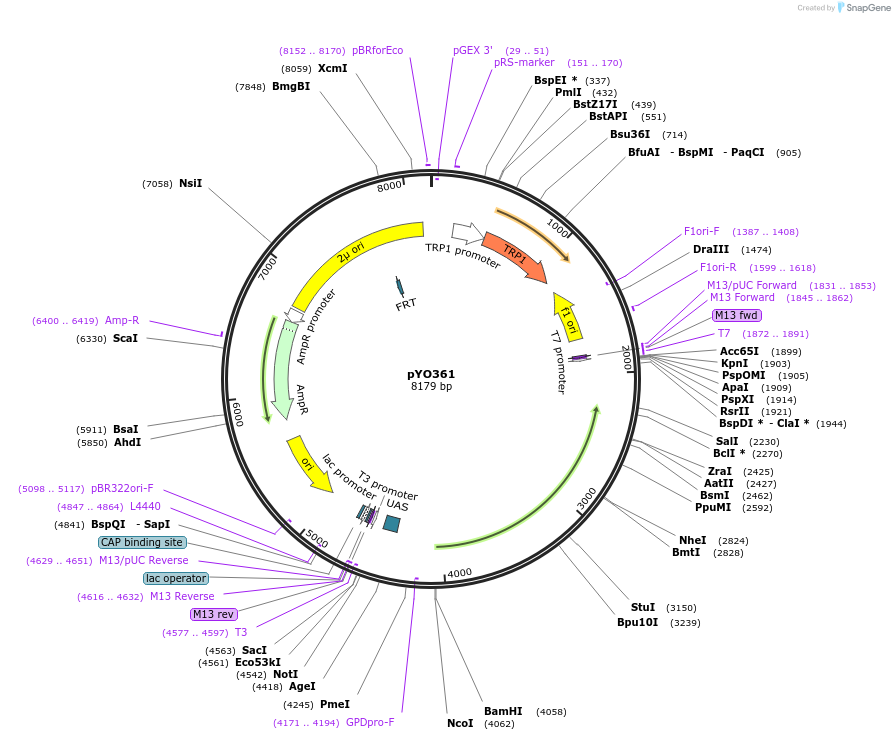
pYO361
Plasmid#235748PurposeProtein expression of ScVPS34 HELCATDepositorInsertVPS34 HELCAT
ExpressionYeastMutationaa 268-875Available SinceMay 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
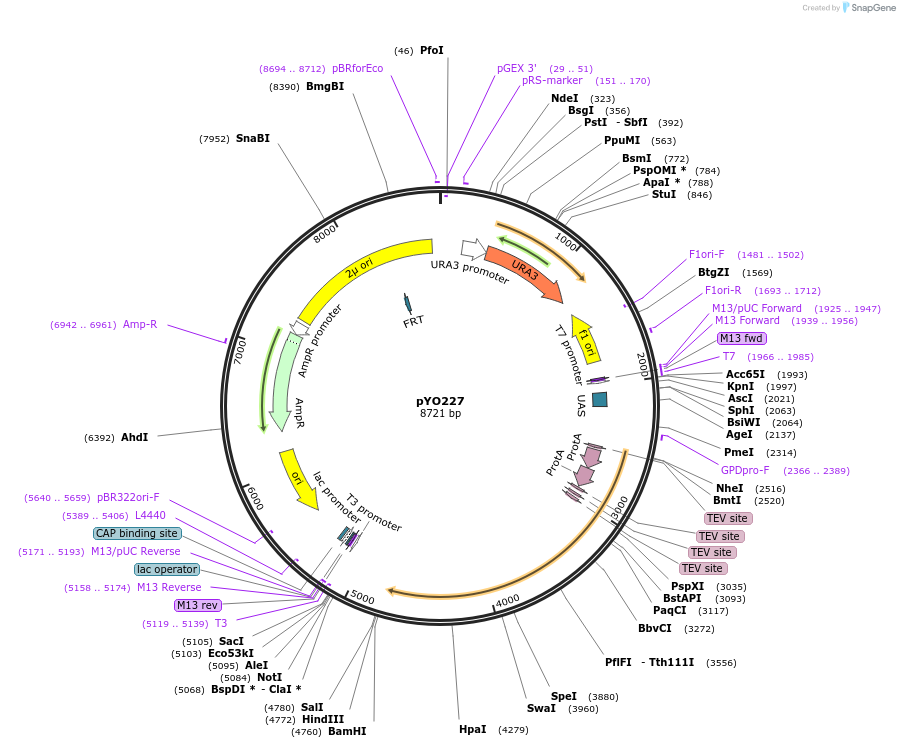
pYO227
Plasmid#235749PurposeProtein expression of ZZ-FL ScVPS30DepositorInsertVPS30
TagsZZ-3xTEVExpressionYeastAvailable SinceMay 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
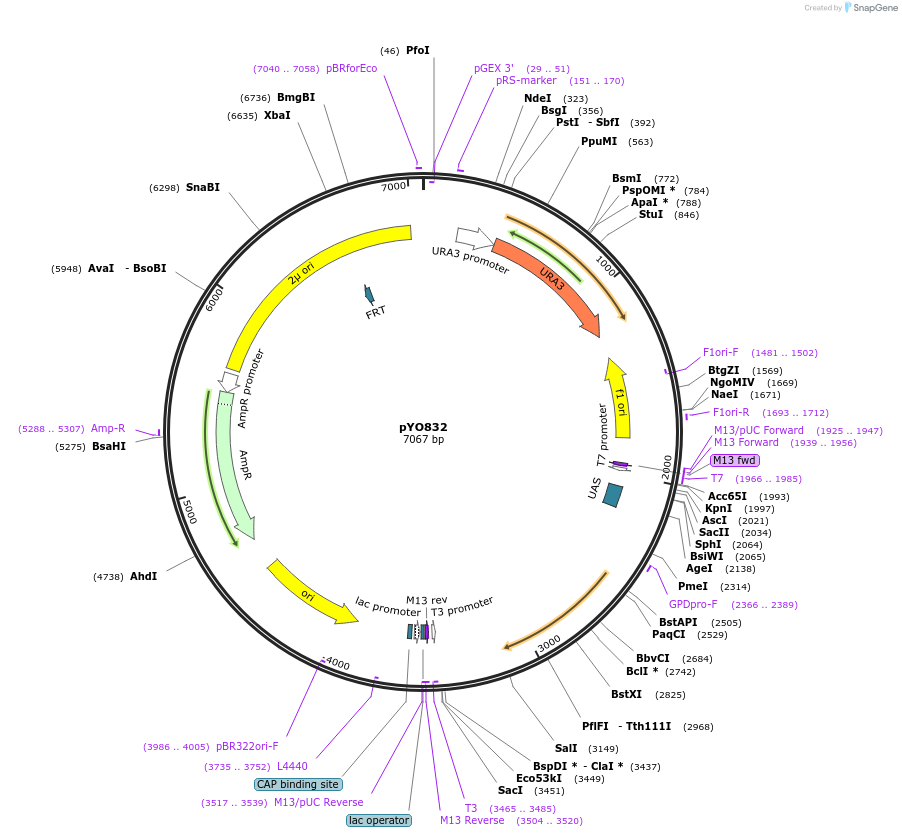
pYO832
Plasmid#235755PurposeProtein expression of ScVPS30 (1-215E), untaggedDepositorInsertScVPS30
ExpressionYeastMutationaa 1-215EAvailable SinceMay 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
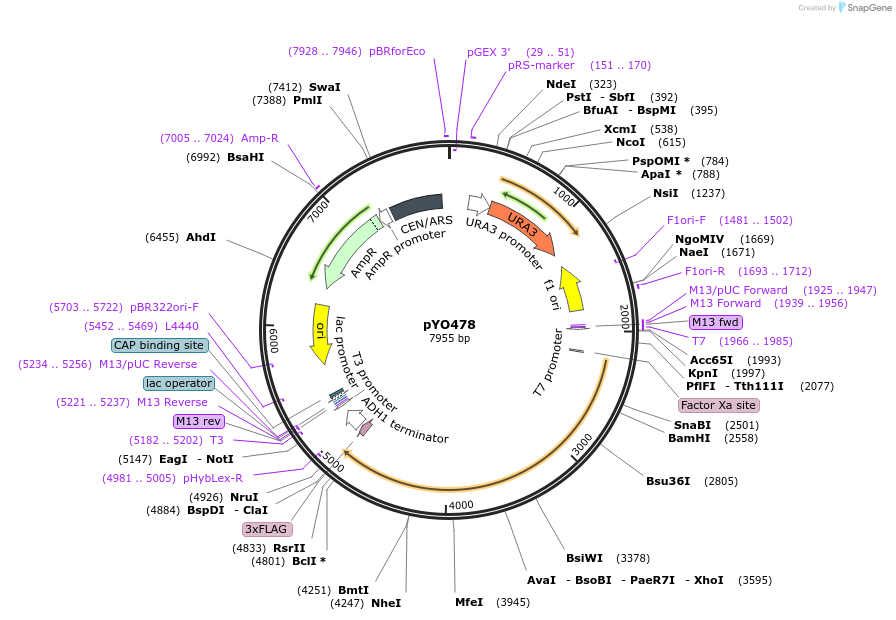
pYO478
Plasmid#235757PurposeExpression of FL ScVPS34-3xFLAG under its own promoterDepositorInsertVPS34
Tags3xFLAGExpressionYeastPromoterScVPS34 promoterAvailable SinceMay 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
pYO764
Plasmid#235761PurposeExpression of ScVPS38-EGFP under its own promoterDepositorInsertVPS38
TagsEGFPExpressionYeastPromoterScVPS38 promoterAvailable SinceMay 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
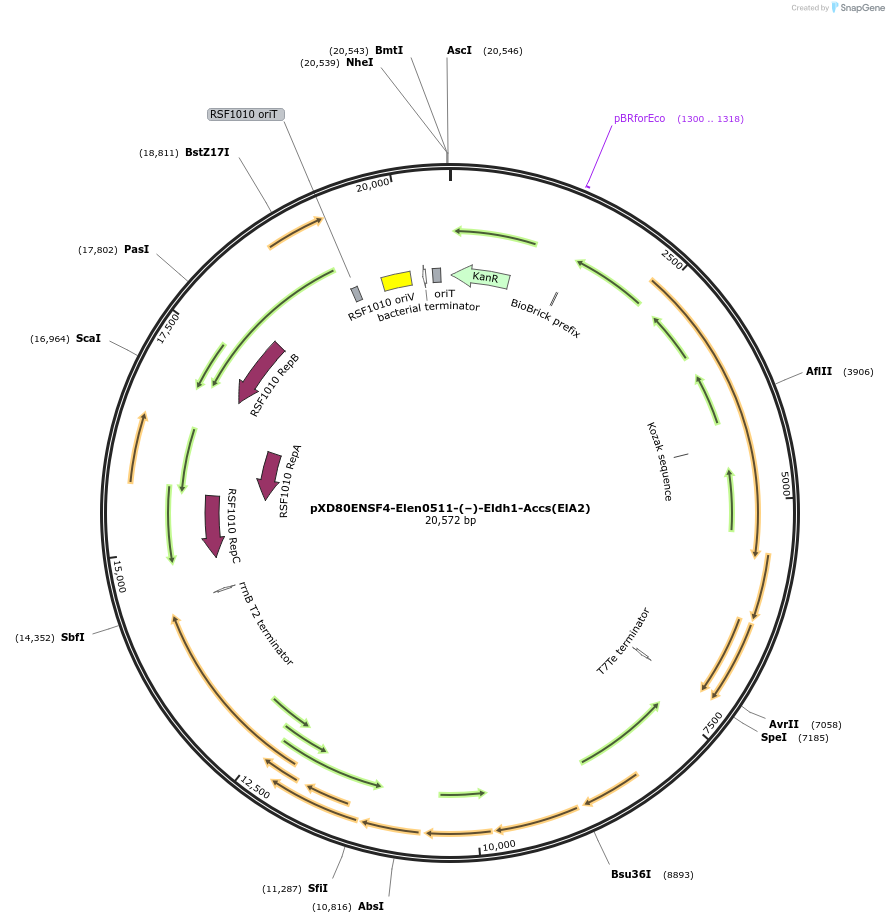
pXD80ENSF4-Elen0511-(–)-Eldh1-Accs(ElA2)
Plasmid#225299PurposeExpresses Eggerthella lenta A2 (–)-Eldh1 in Gordonibacter urolithinfaciens DSM 27213DepositorInsert(–)-Eldh1
ExpressionBacterialAvailable SinceMay 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
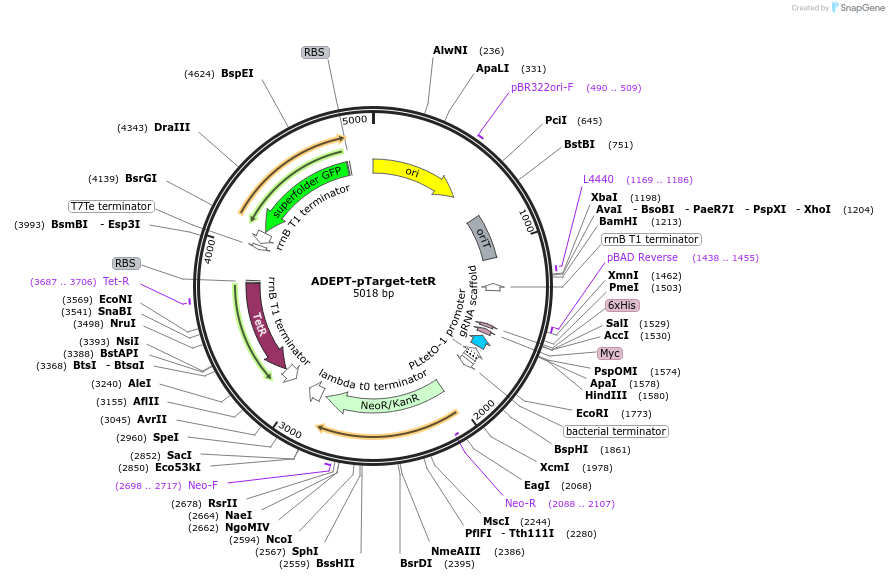
ADEPT-pTarget-tetR
Plasmid#238043PurposeModified ADEPT-pCas9 with sfGFP under TtrB promoter for tetrathionate (TTR) sensing.DepositorInsertsfGFP
UseSynthetic BiologyPromoterttrBAvailable SinceMay 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
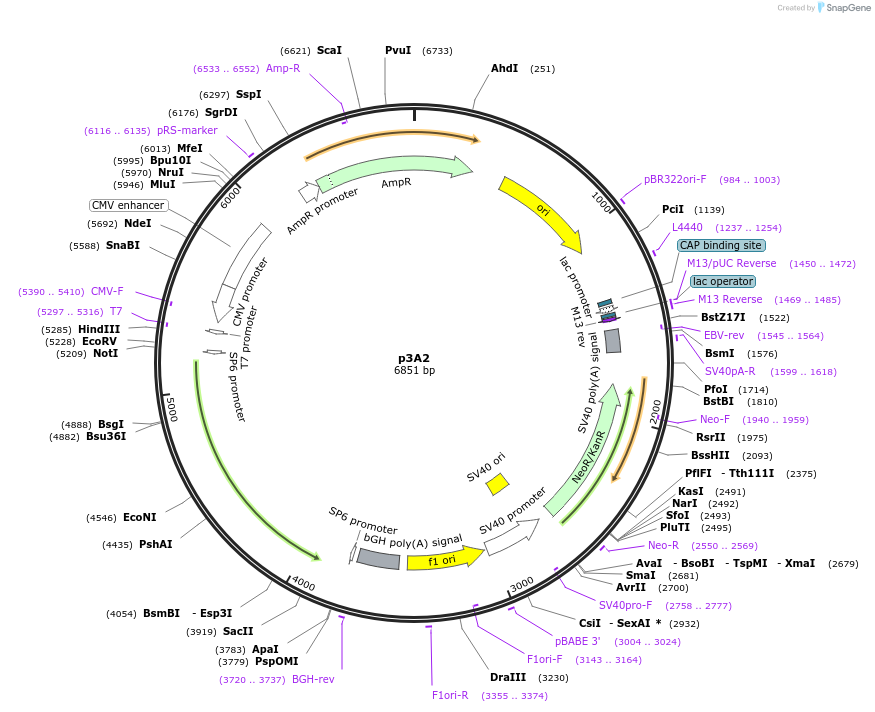
p3A2
Plasmid#234793PurposeMammalian expression of bovine arrestin-2DepositorAvailable SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
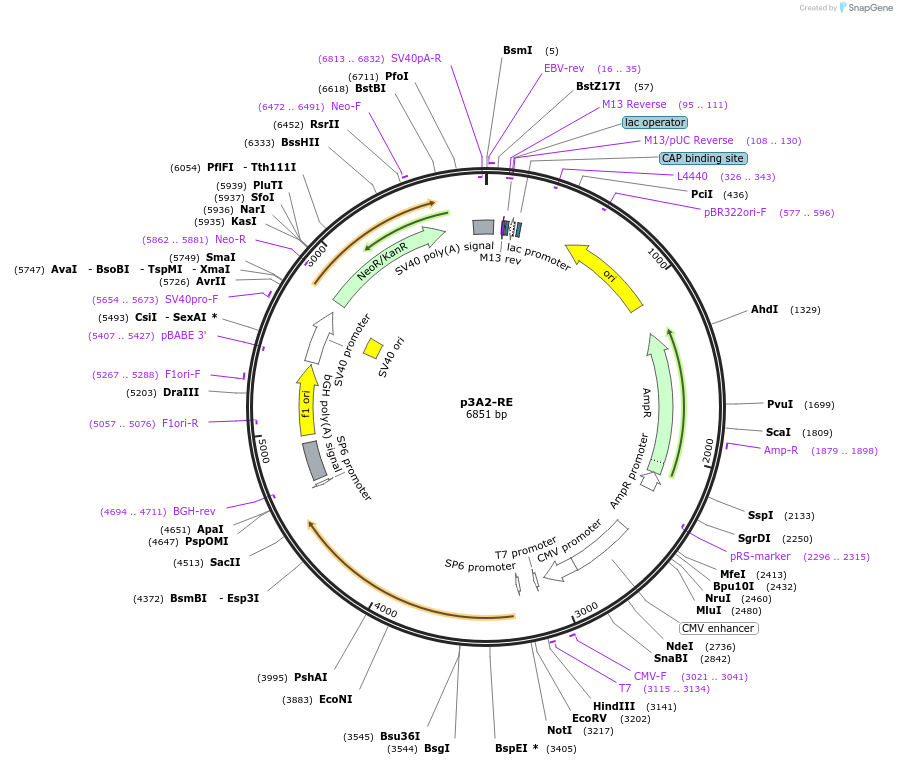
p3A2-RE
Plasmid#234794PurposeMammalian expression of bovine arrestin-2 (R169E)DepositorAvailable SinceApril 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
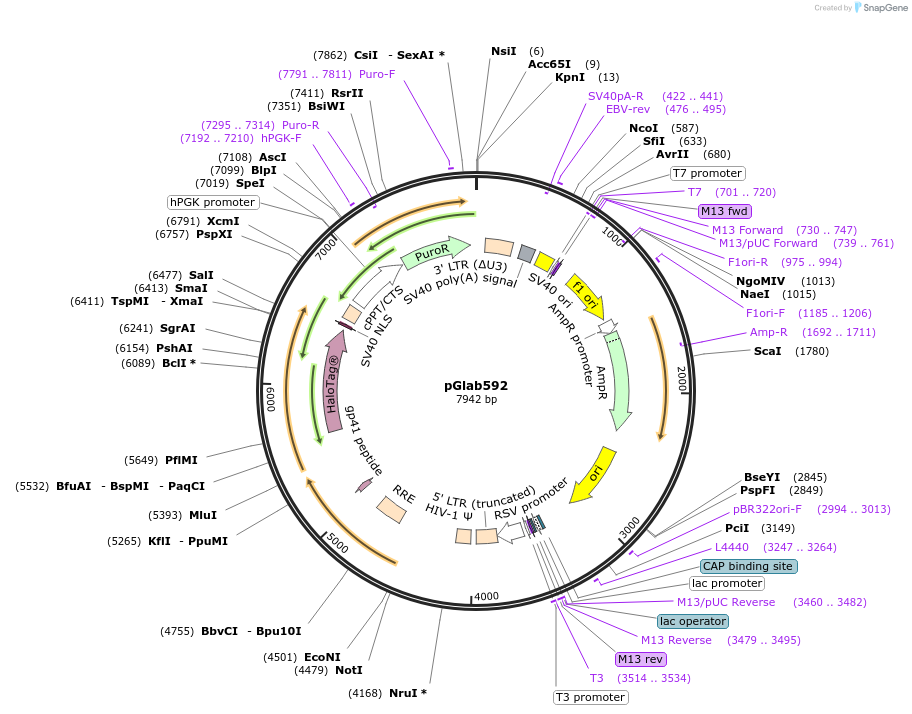
pGlab592
Plasmid#236554Purpose240 basepair upstream sequence of KSHV ORF68 driving HaloTag expressionDepositorInsertKSHV ORF68 upstream 240bp
UseLentiviralTagsHaloTagExpressionMammalianAvailable SinceApril 15, 2025AvailabilityAcademic Institutions and Nonprofits only -
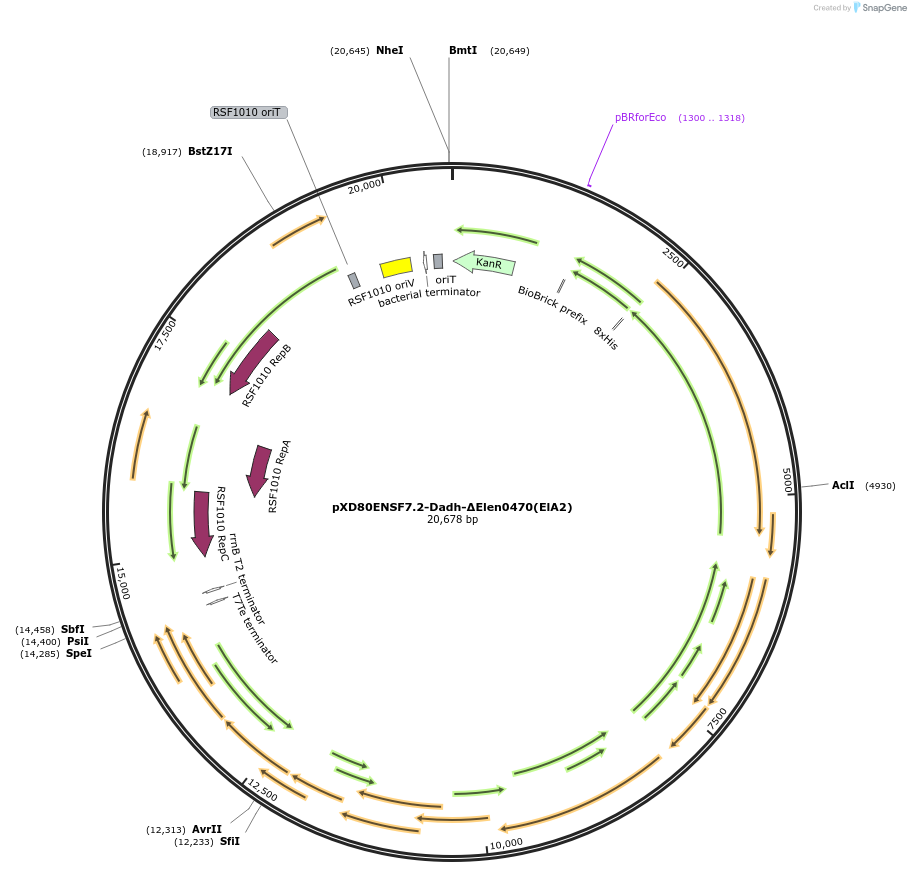
pXD80ENSF7.2-Dadh-ΔElen0470(ElA2)
Plasmid#225317PurposeExpresses Eggerthella lenta A2 Dadh (ΔElen_0470) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertDadh (ΔElen_0470)
TagsHis8ExpressionBacterialAvailable SinceApril 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
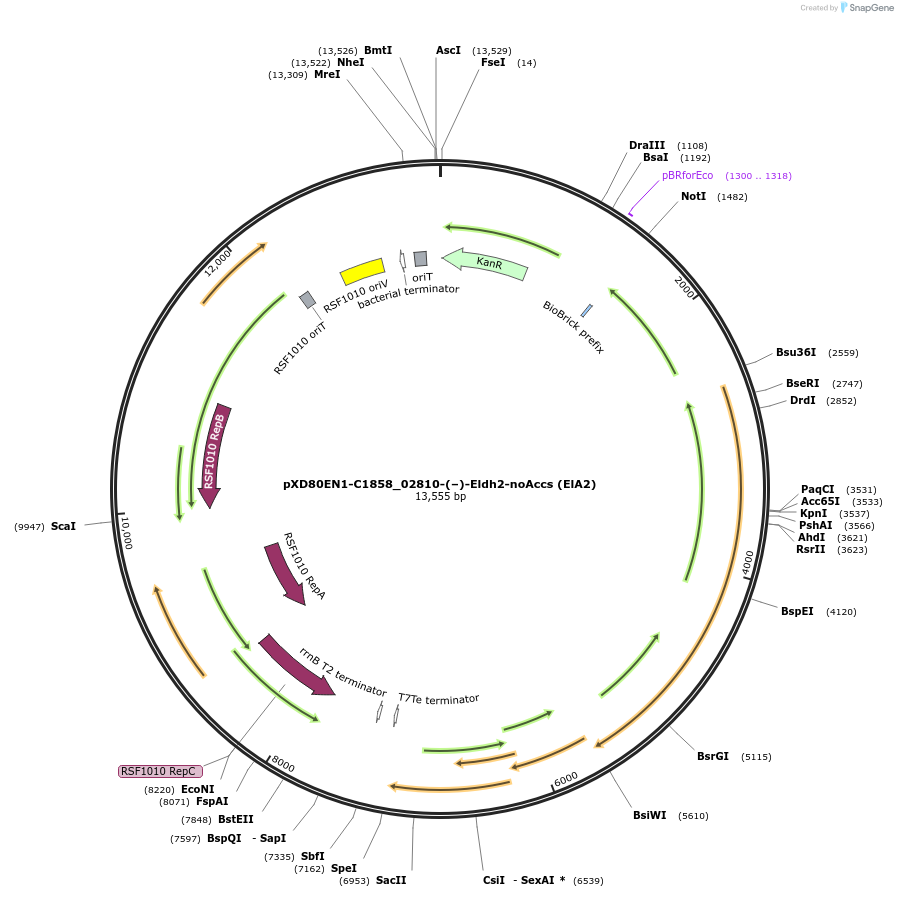
pXD80EN1-C1858_02810-(–)-Eldh2-noAccs (ElA2)
Plasmid#225309PurposeExpresses Eggerthella lenta A2 (–)-Eldh2 without accessory genes (inactive) in Gordonibacter urolithinfaciens DSM 27213DepositorInsert(–)-Eldh2
ExpressionBacterialAvailable SinceApril 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
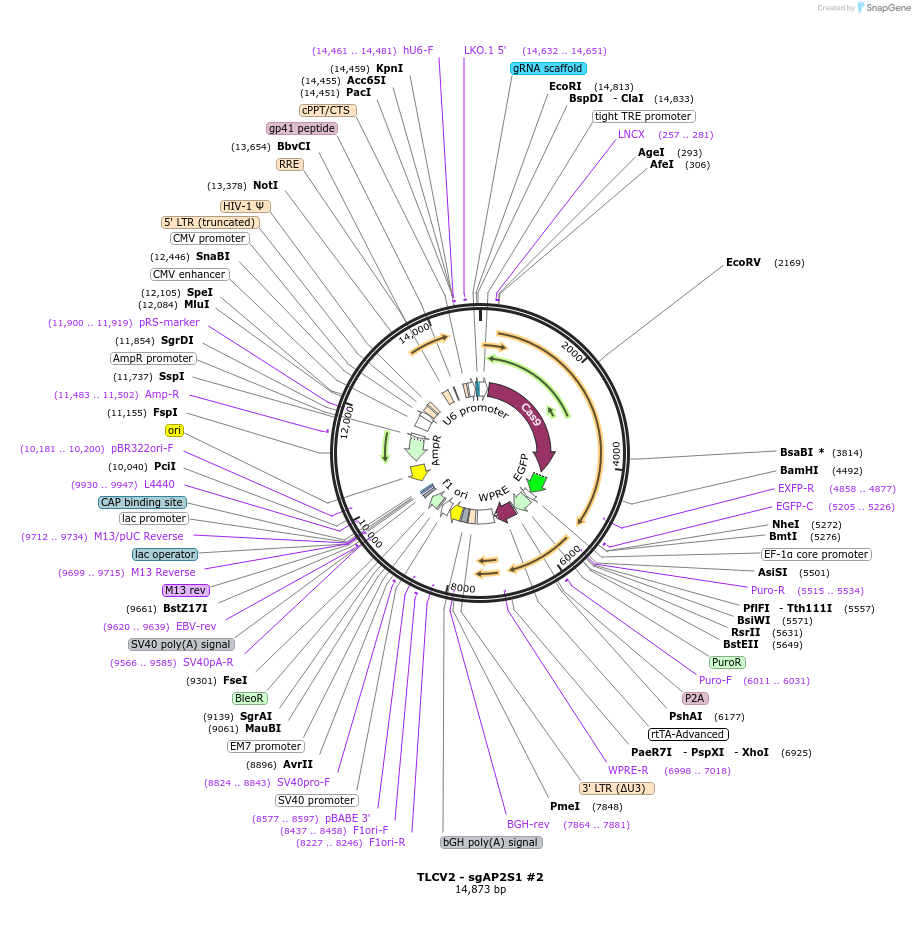
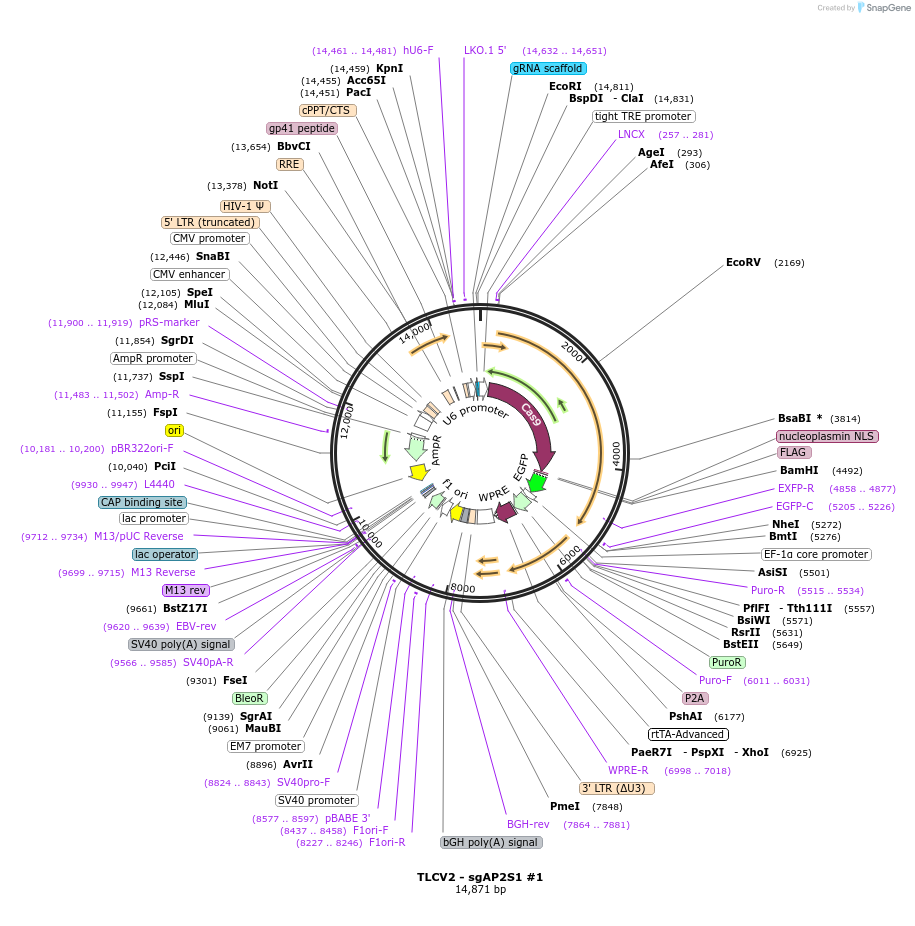
TLCV2 - sgAP2S1 #2
Plasmid#202757PurposeEncodes Doxycycline inducible Cas9 and sgRNA targeting AP2S1DepositorInsertgRNA targeting AP2S1
UseCRISPR and LentiviralExpressionMammalianAvailable SinceApril 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
TLCV2 - sgAP2S1 #1
Plasmid#202756PurposeEncodes Doxycycline inducible Cas9 and sgRNA targeting AP2S1DepositorInsertgRNA targeting AP2S1
UseCRISPR and LentiviralExpressionMammalianAvailable SinceApril 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
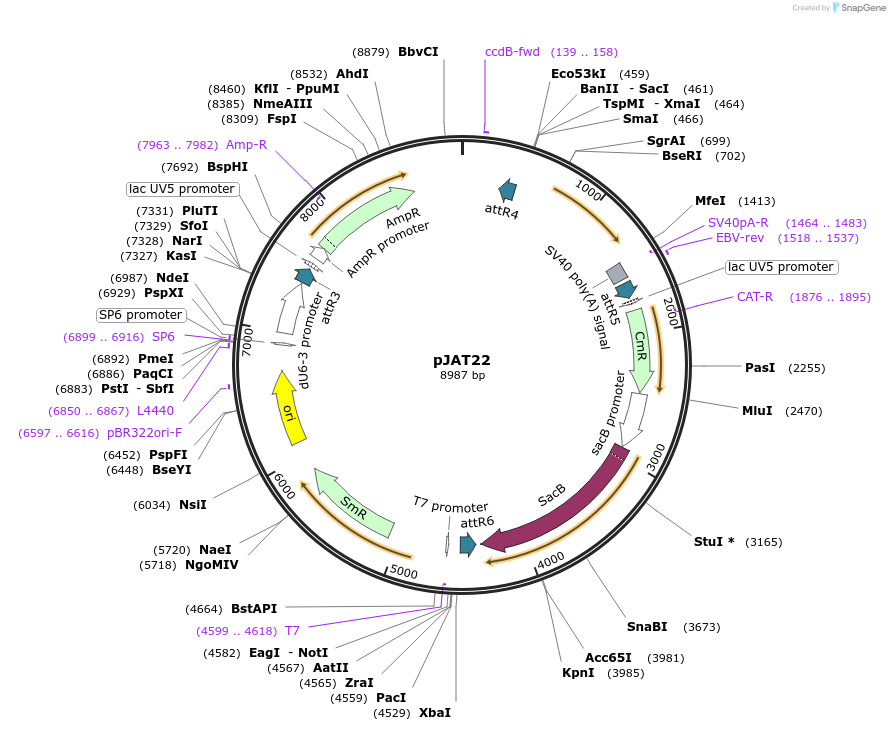
pJAT22
Plasmid#204311PurposeFor generating targeting inversions. N-terminal split GFP, expressed in eyes.DepositorInsert3XP3-GFPn
UseCRISPRPromoter3XP3Available SinceMarch 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
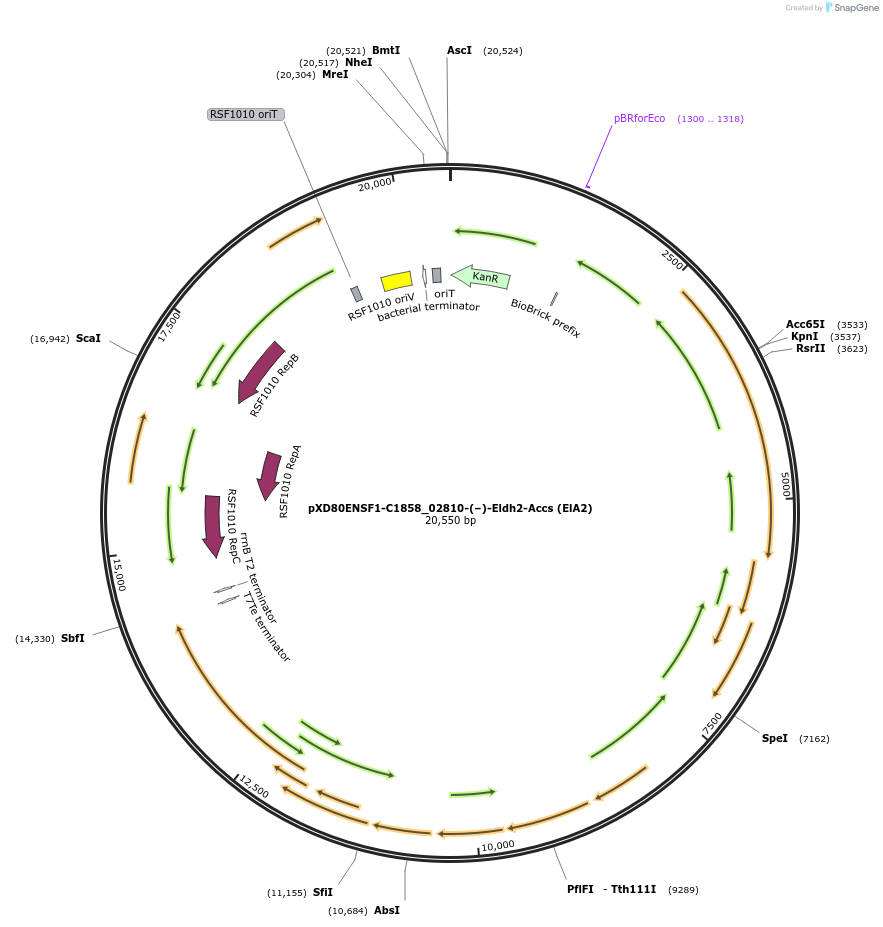
pXD80ENSF1-C1858_02810-(–)-Eldh2-Accs (ElA2)
Plasmid#225296PurposeExpresses Eggerthella lenta A2 (–)-Eldh2 in Gordonibacter urolithinfaciens DSM 27213DepositorInsert(–)-Eldh2
ExpressionBacterialAvailable SinceMarch 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
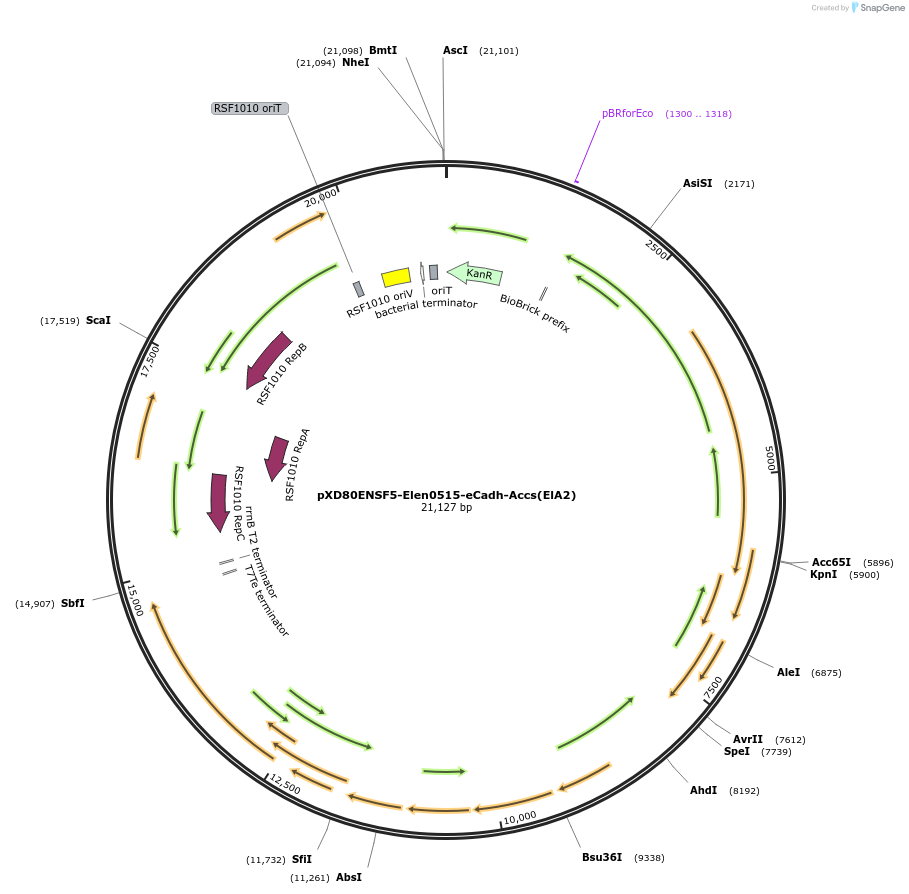
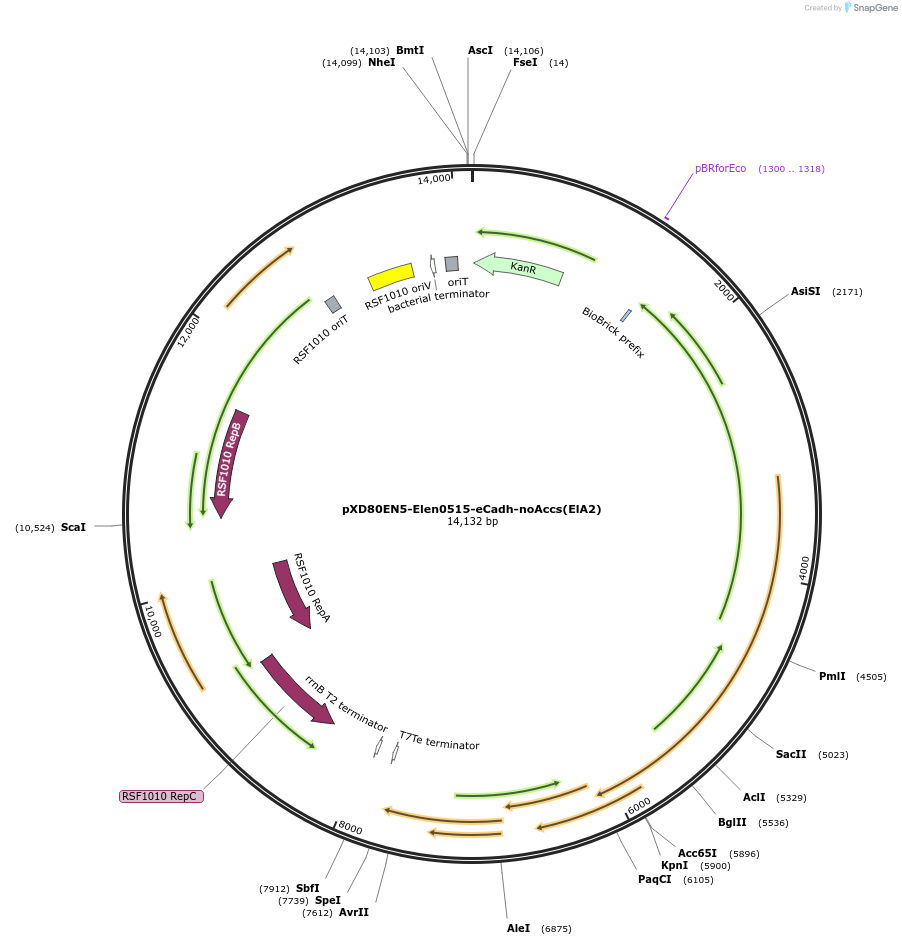
pXD80ENSF5-Elen0515-eCadh-Accs(ElA2)
Plasmid#225300PurposeExpresses Eggerthella lenta A2 eCadh in Gordonibacter urolithinfaciens DSM 27213DepositorInserteCadh
ExpressionBacterialAvailable SinceMarch 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
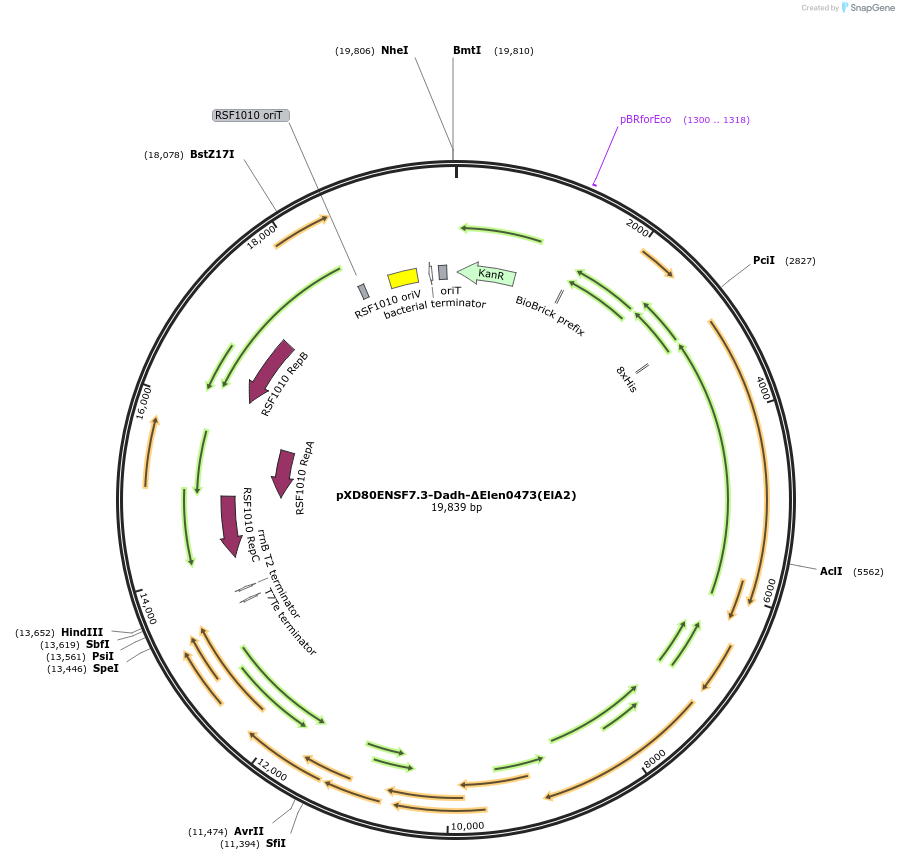
pXD80ENSF7.3-Dadh-ΔElen0473(ElA2)
Plasmid#225318PurposeExpresses Eggerthella lenta A2 Dadh (ΔElen_0473) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertDadh (ΔElen_0473)
TagsHis8ExpressionBacterialAvailable SinceMarch 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
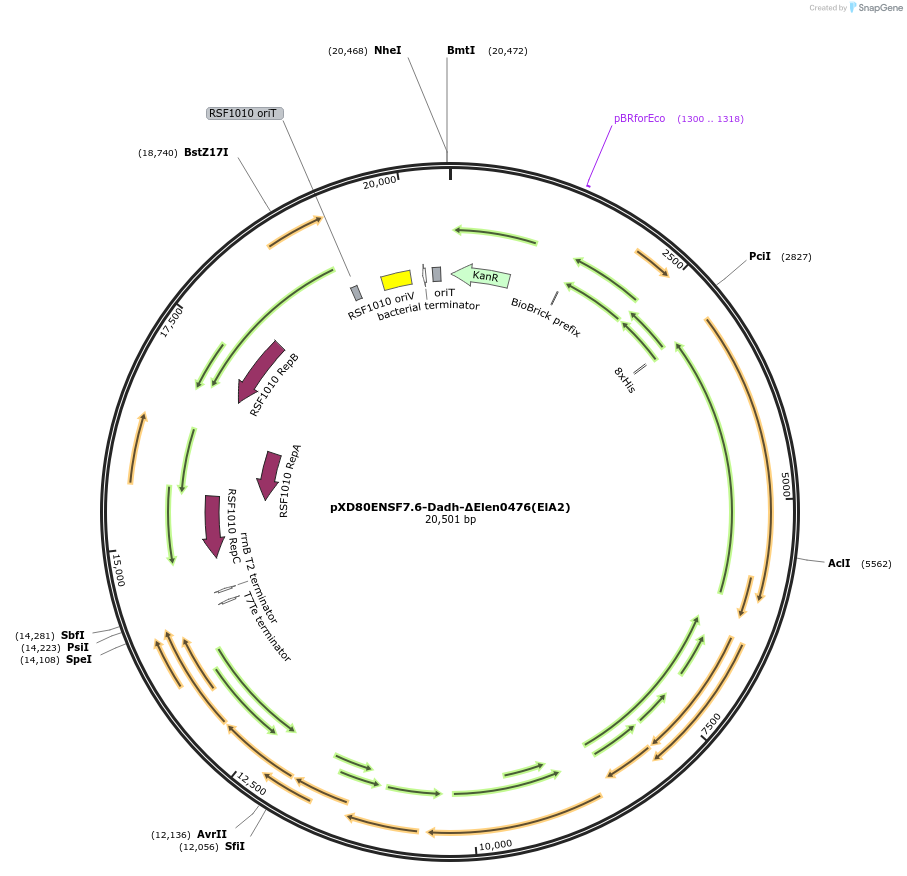
pXD80ENSF7.6-Dadh-ΔElen0476(ElA2)
Plasmid#225321PurposeExpresses Eggerthella lenta A2 Dadh (ΔElen_0476) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertDadh (ΔElen_0476)
TagsHis8ExpressionBacterialAvailable SinceMarch 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
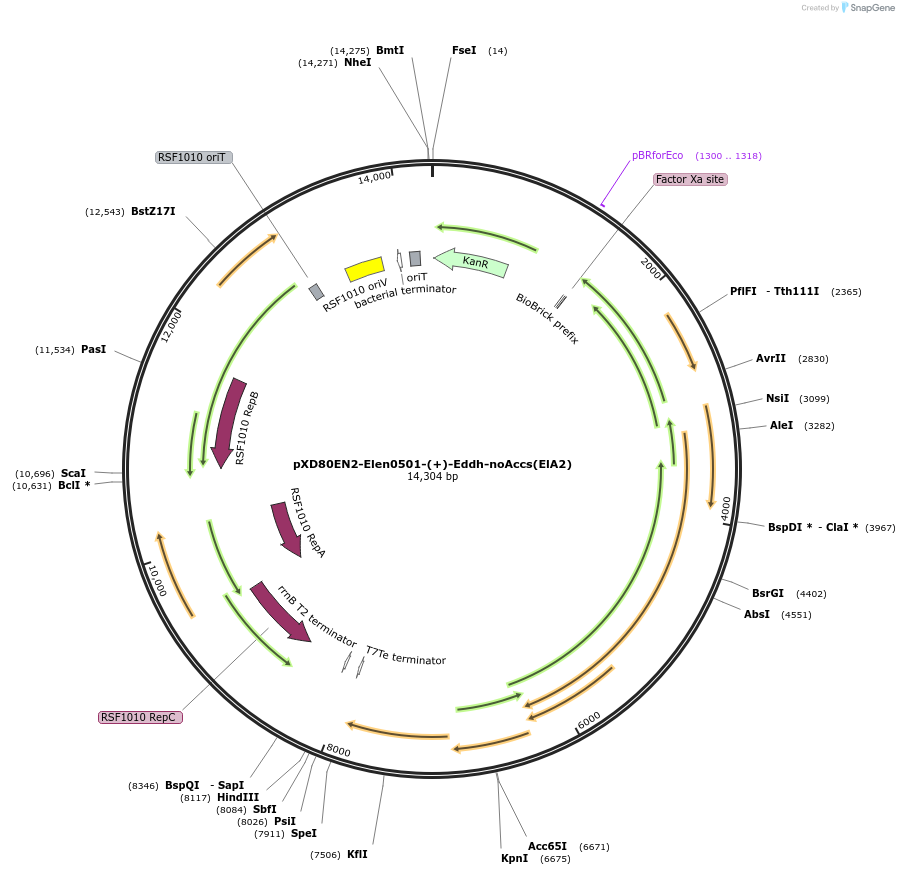
pXD80EN2-Elen0501-(+)-Eddh-noAccs(ElA2)
Plasmid#225310PurposeExpresses Eggerthella lenta A2 (+)-Eddh without accessory genes (inactive) in Gordonibacter urolithinfaciens DSM 27213DepositorInsert(+)-Eddh
ExpressionBacterialAvailable SinceMarch 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
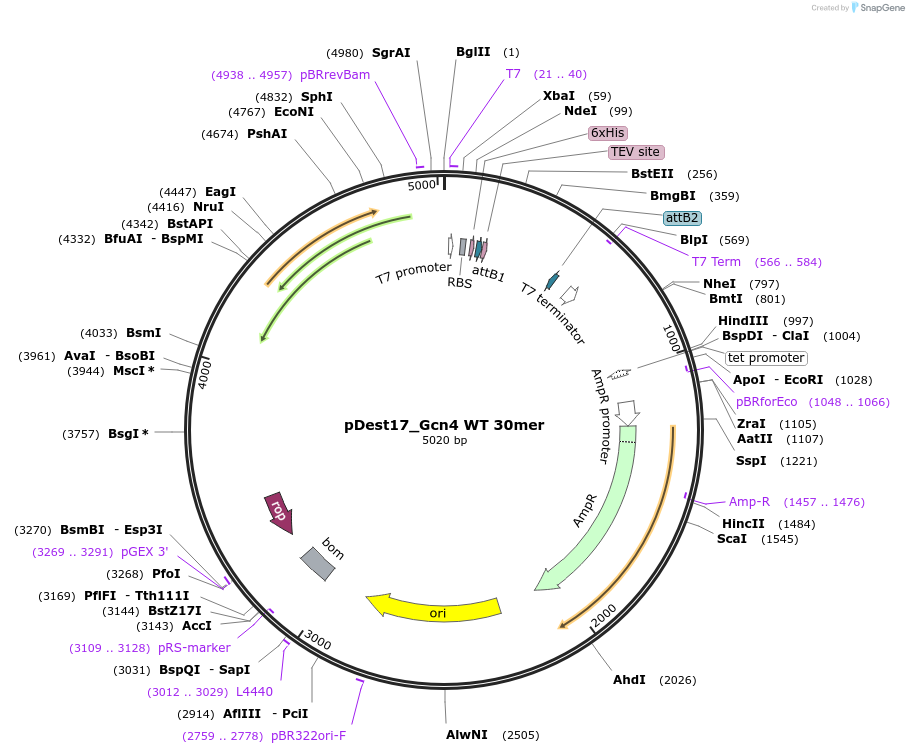
pDest17_Gcn4 WT 30mer
Plasmid#231888PurposeBacterial expression of N-terminally 6His tagged Gcn4 WT 30merDepositorInsertGcn4 30mer
Tags6xHis, TEV cleavage siteExpressionBacterialPromoterT7Available SinceMarch 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
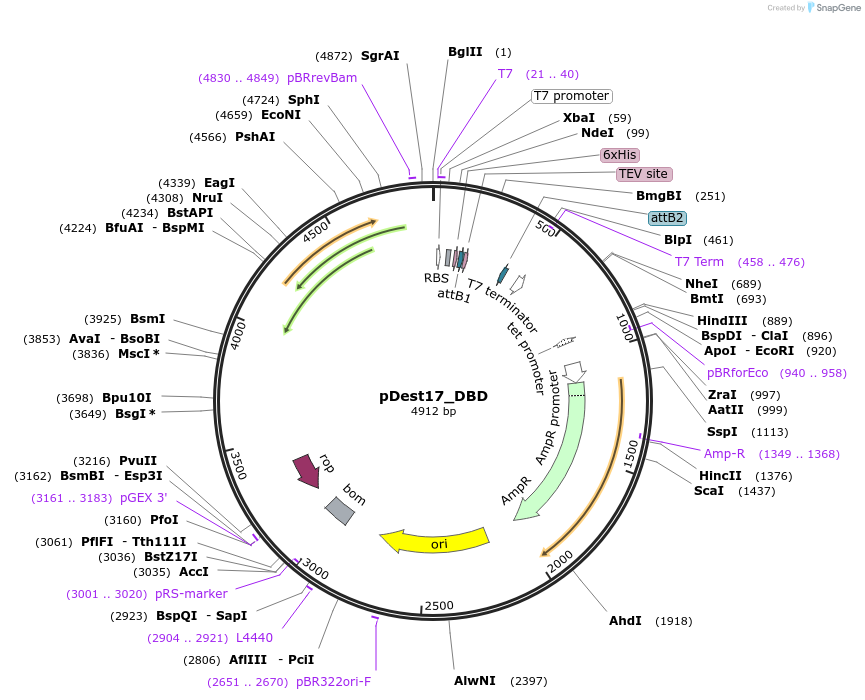
pDest17_DBD
Plasmid#231886PurposeBacterial expression of N-terminally 6His tagged Gcn4 DBDDepositorInsertGcn4 DBD
Tags6xHis, TEV cleavage siteExpressionBacterialPromoterT7Available SinceMarch 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
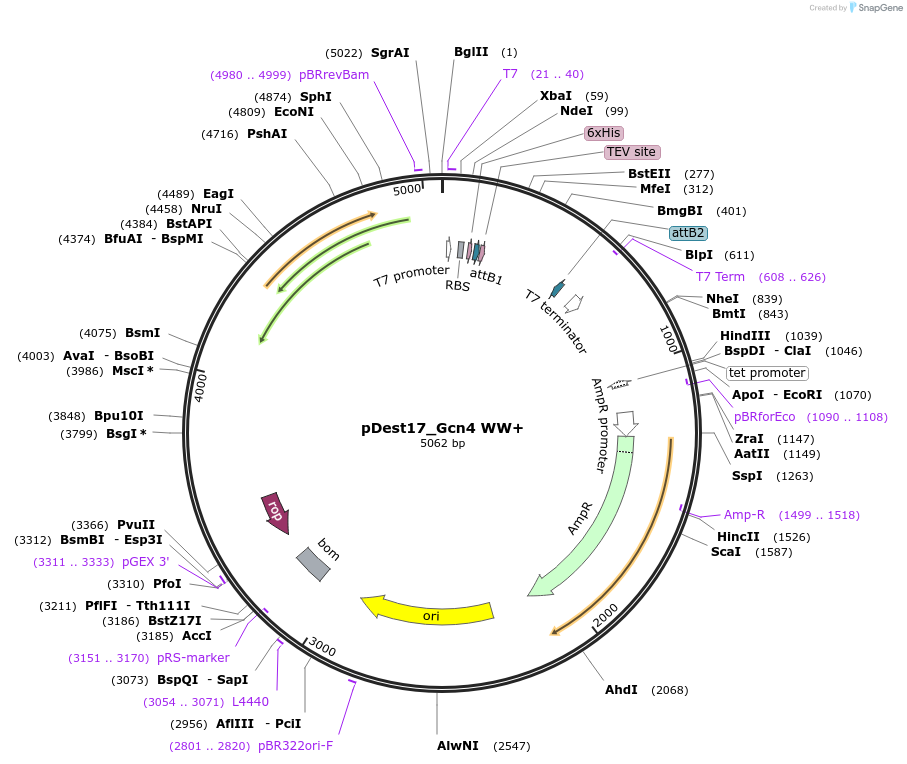
pDest17_Gcn4 WW+
Plasmid#231885PurposeBacterial expression of N-terminally 6His tagged Gcn4 WW+DepositorInsertGcn4 WW+
Tags6xHis, TEV cleavage siteExpressionBacterialMutationE109N, S117D, T121W, N126W, T132WPromoterT7Available SinceMarch 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
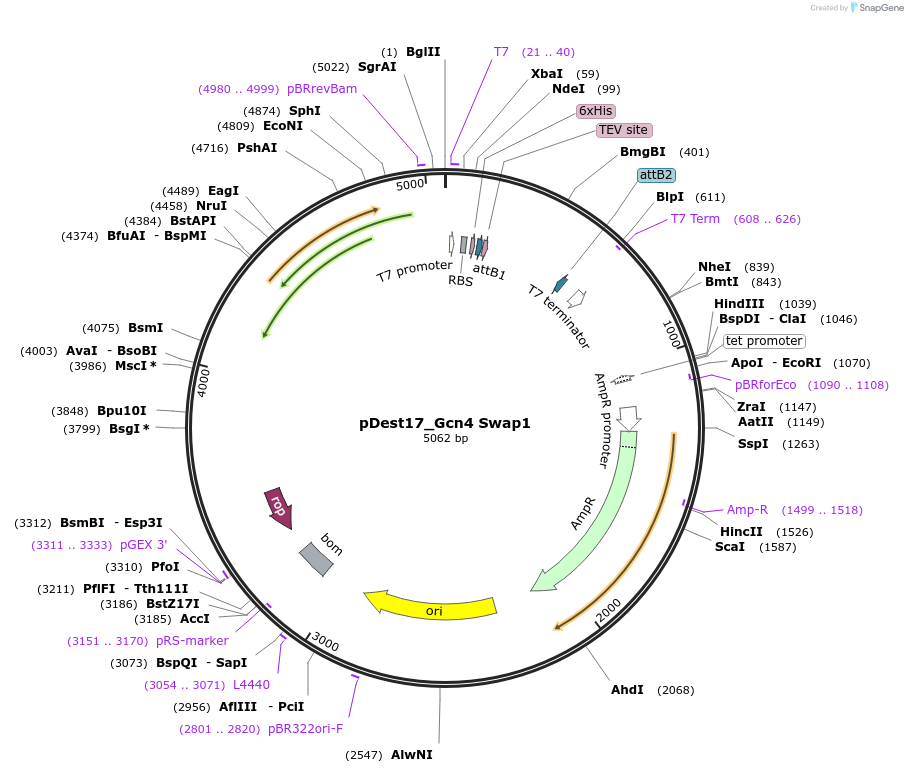
pDest17_Gcn4 Swap1
Plasmid#231879PurposeBacterial expression of N-terminally 6His tagged Gcn4 Swap1DepositorInsertGcn4 Swap1
Tags6xHis, TEV cleavage siteExpressionBacterialPromoterT7Available SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
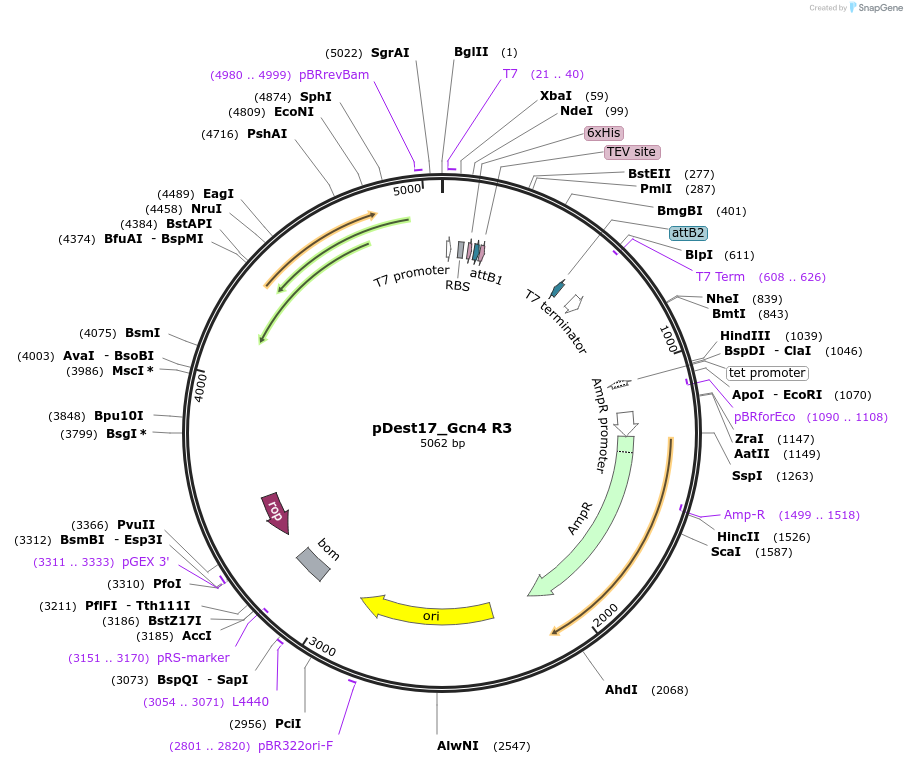
pDest17_Gcn4 R3
Plasmid#231874PurposeBacterial expression of N-terminally 6His tagged Gcn4 R3DepositorInsertGcn4 R3
Tags6xHis, TEV cleavage siteExpressionBacterialMutationD103R, D133R, K143RPromoterT7Available SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
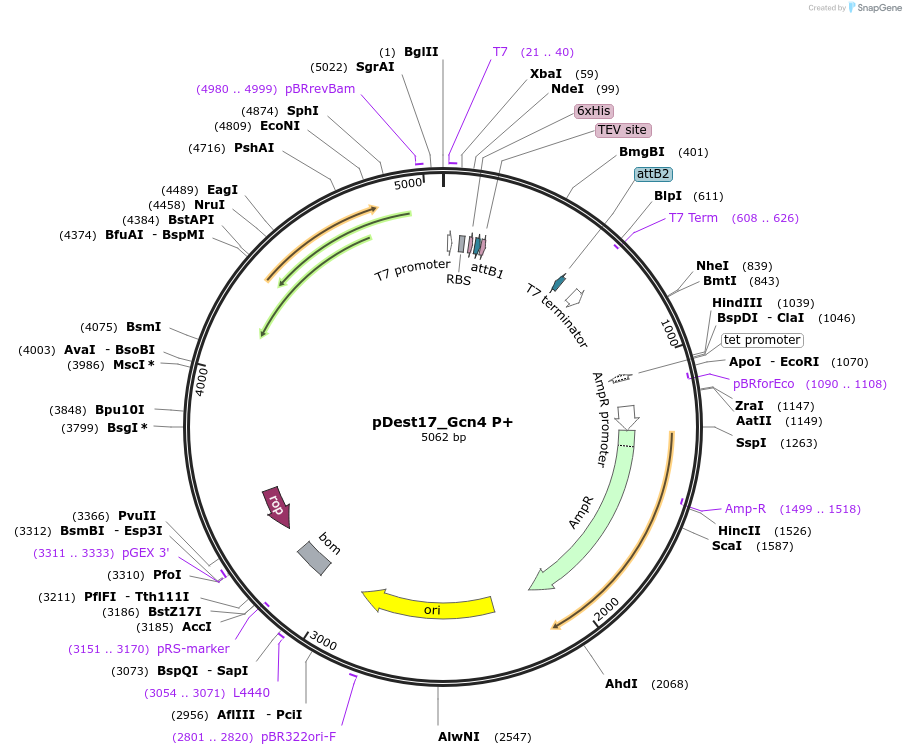
pDest17_Gcn4 P+
Plasmid#231872PurposeBacterial expression of N-terminally 6His tagged Gcn4 P+DepositorInsertGcn4 P+
Tags6xHis, TEV cleavage siteExpressionBacterialMutationS122P, T131F, A141P, E143DPromoterT7Available SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
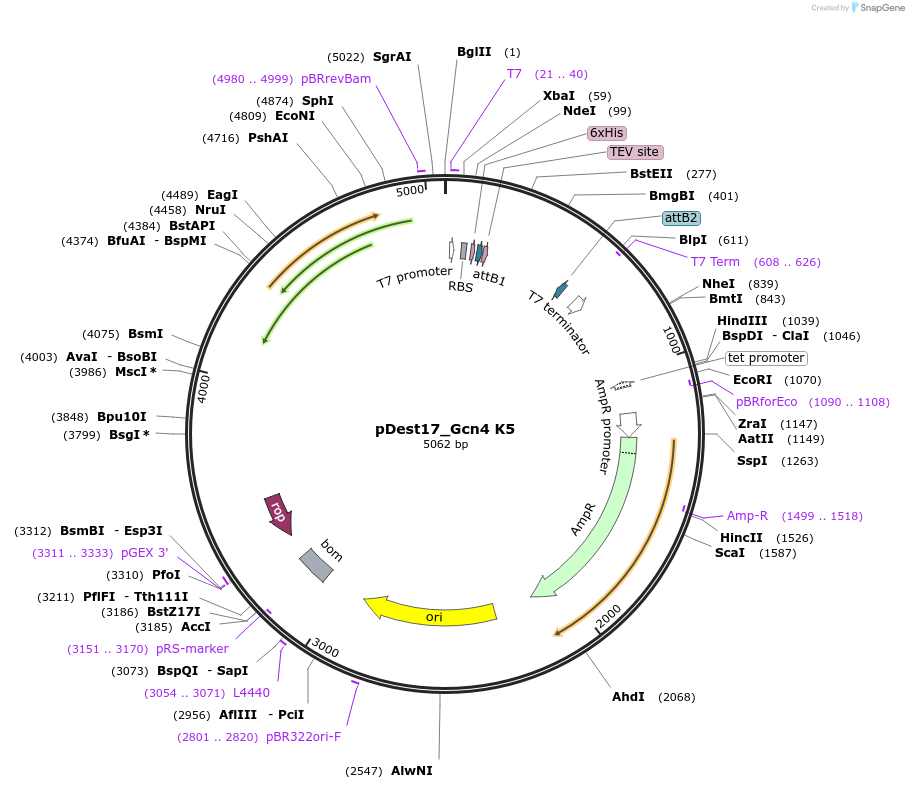
pDest17_Gcn4 K5
Plasmid#231866PurposeBacterial expression of N-terminally 6His tagged Gcn4 K5DepositorInsertGcn4 K5
Tags6xHis, TEV cleavage siteExpressionBacterialMutationD103K, D115K, D127K, D133K, E143KPromoterT7Available SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
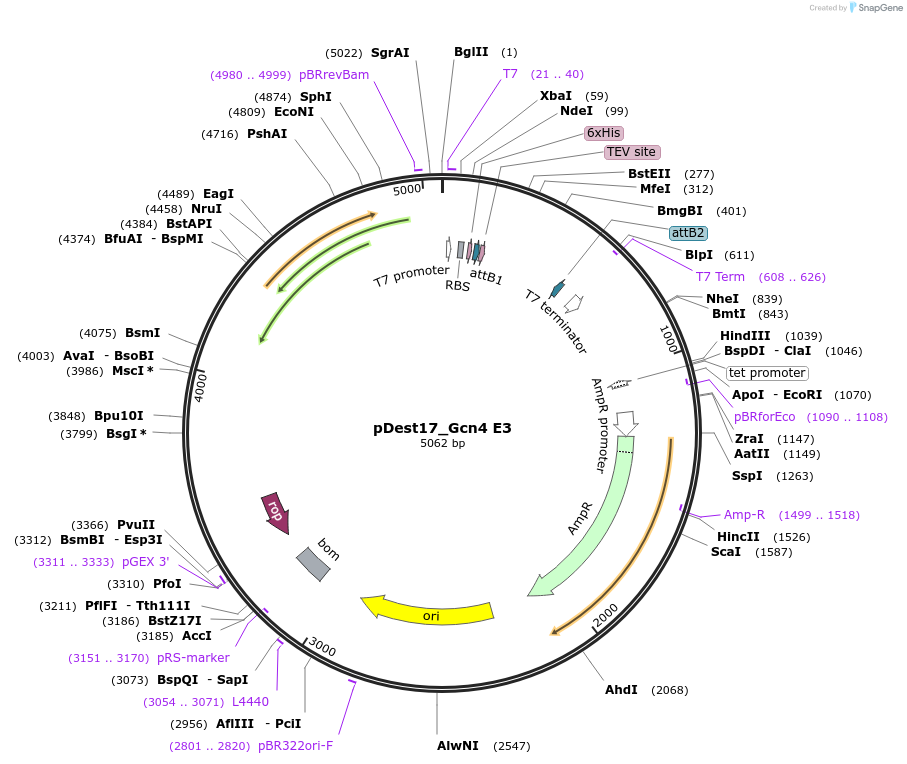
pDest17_Gcn4 E3
Plasmid#231863PurposeBacterial expression of N-terminally 6His tagged Gcn4 E3DepositorInsertGcn4 E3
Tags6xHis, TEV cleavage siteExpressionBacterialMutationD105E, D118E, D139EPromoterT7Available SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
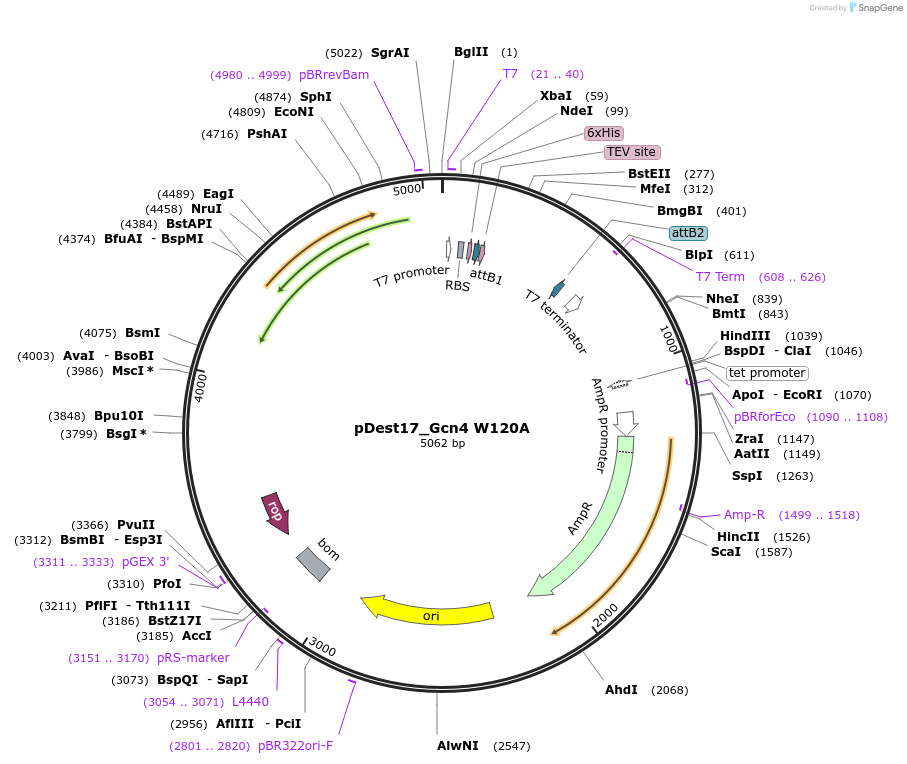
pDest17_Gcn4 W120A
Plasmid#231862PurposeBacterial expression of N-terminally 6His tagged Gcn4 W120ADepositorInsertGcn4
Tags6xHis, TEV cleavage siteExpressionBacterialMutationW120APromoterT7Available SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
pXD80EN5-Elen0515-eCadh-noAccs(ElA2)
Plasmid#225313PurposeExpresses Eggerthella lenta A2 eCadh without accessory genes (inactive) in Gordonibacter urolithinfaciens DSM 27213DepositorInserteCadh (Elen_0515 )
ExpressionBacterialAvailable SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
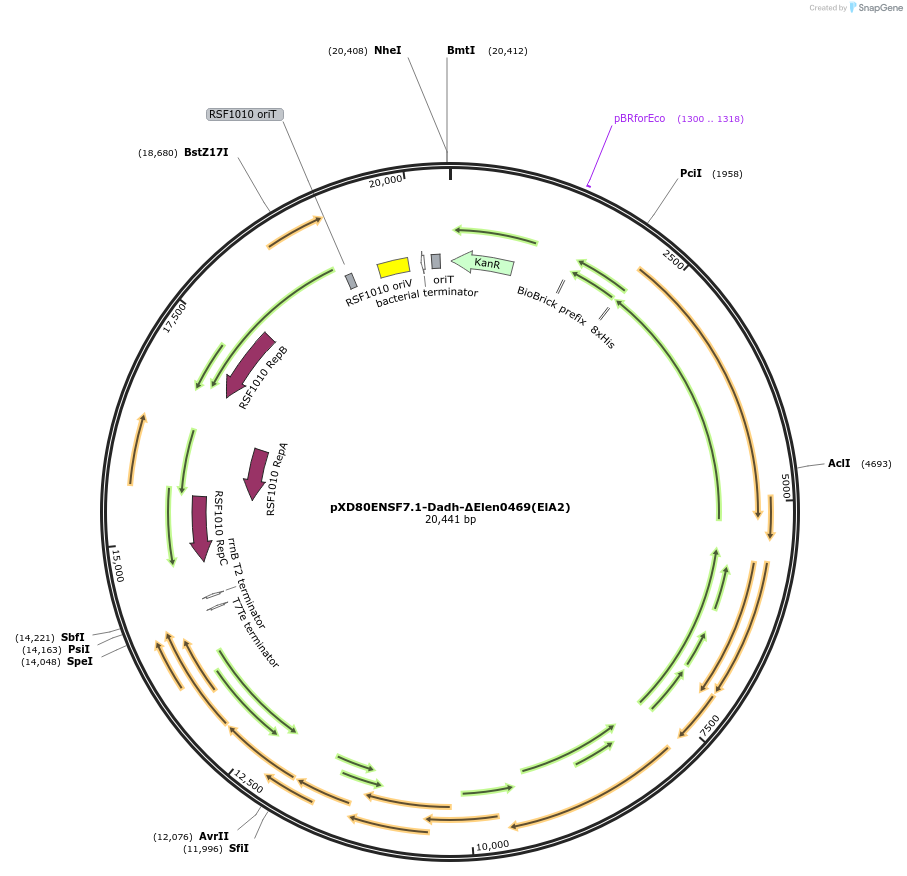
pXD80ENSF7.1-Dadh-ΔElen0469(ElA2)
Plasmid#225315PurposeExpresses Eggerthella lenta A2 Dadh (ΔElen_0469) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertDadh (ΔElen_0469)
TagsHis8ExpressionBacterialAvailable SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
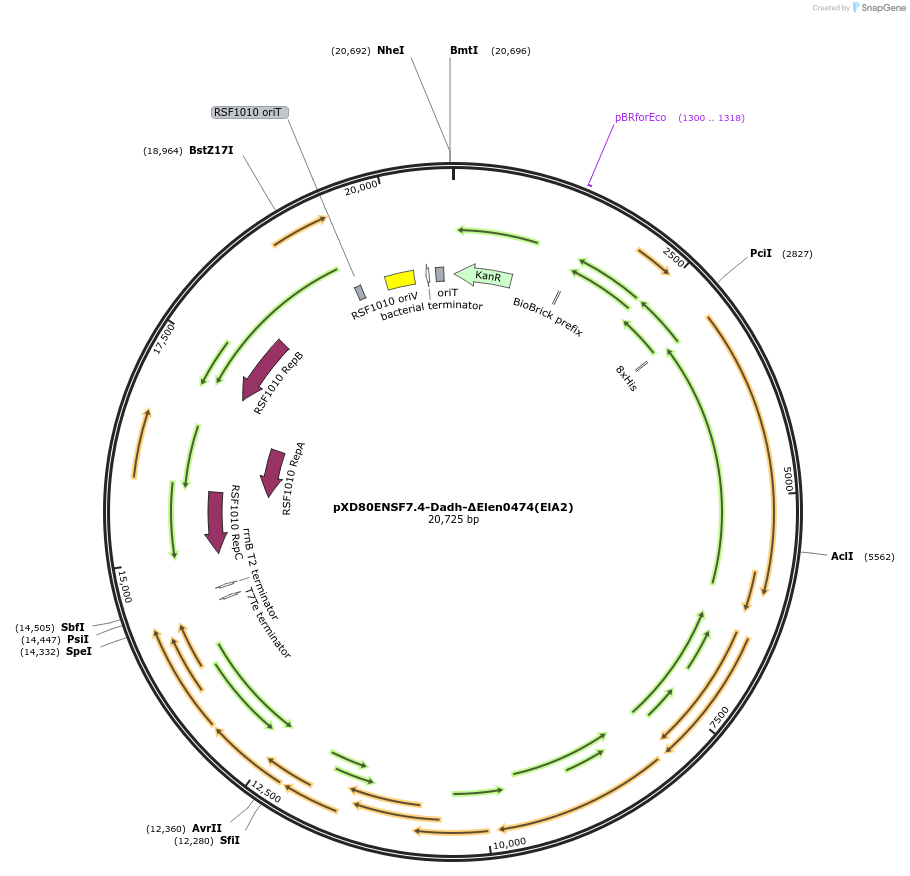
pXD80ENSF7.4-Dadh-ΔElen0474(ElA2)
Plasmid#225319PurposeExpresses Eggerthella lenta A2 Dadh (ΔElen_0474) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertDadh (ΔElen_0474)
TagsHis8ExpressionBacterialAvailable SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
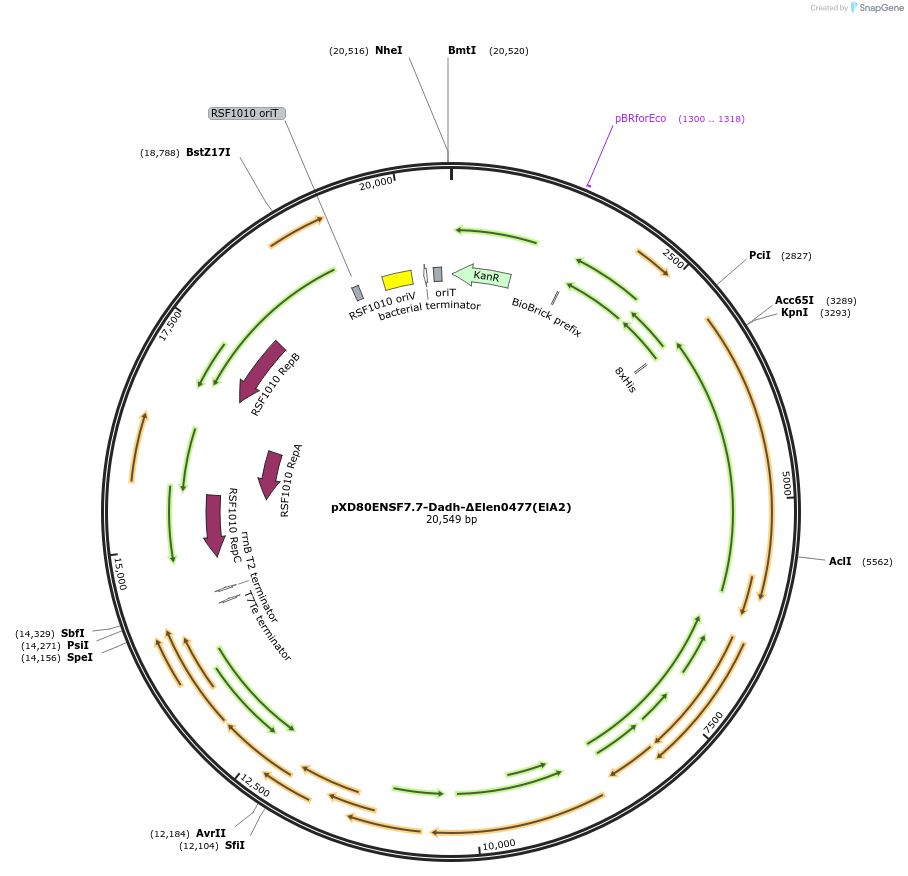
pXD80ENSF7.7-Dadh-ΔElen0477(ElA2)
Plasmid#225322PurposeExpresses Eggerthella lenta A2 Dadh (ΔElen_0477) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertDadh (ΔElen_0477)
TagsHis8ExpressionBacterialAvailable SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
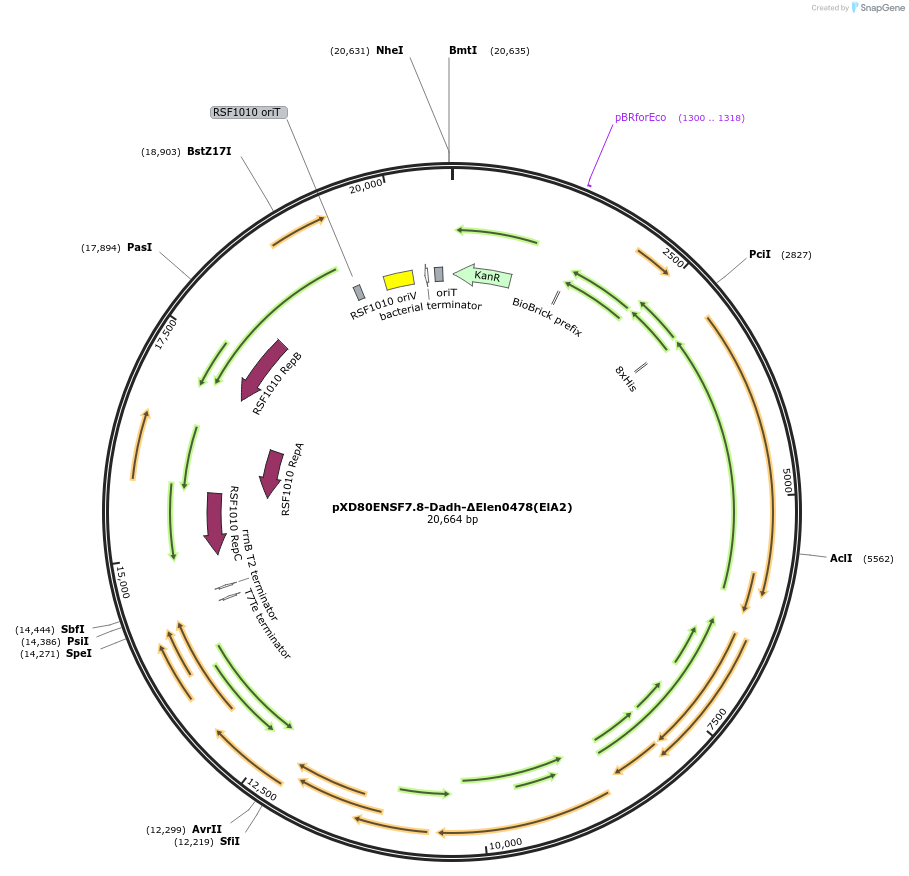
pXD80ENSF7.8-Dadh-ΔElen0478(ElA2)
Plasmid#225323PurposeExpresses Eggerthella lenta A2 Dadh (ΔElen_0478) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertDadh (ΔElen_0478)
TagsHis8ExpressionBacterialAvailable SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
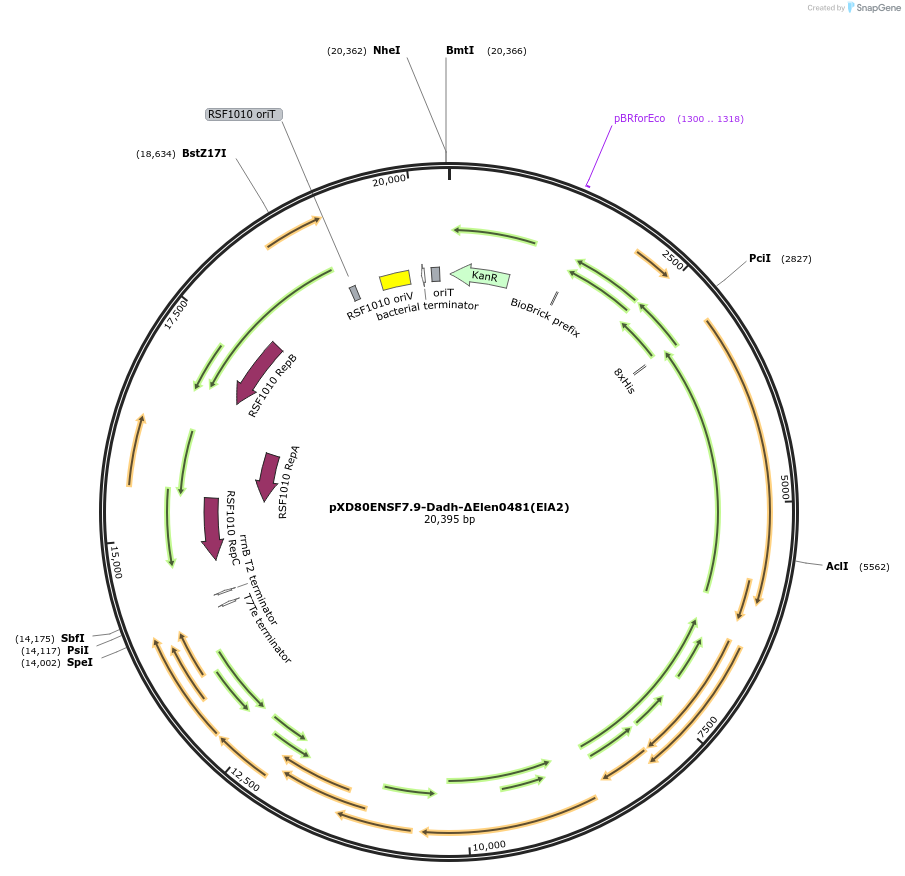
pXD80ENSF7.9-Dadh-ΔElen0481(ElA2)
Plasmid#225324PurposeExpresses Eggerthella lenta A2 Dadh (ΔElen_0481) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertDadh (ΔElen_0481)
TagsHis8ExpressionBacterialAvailable SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
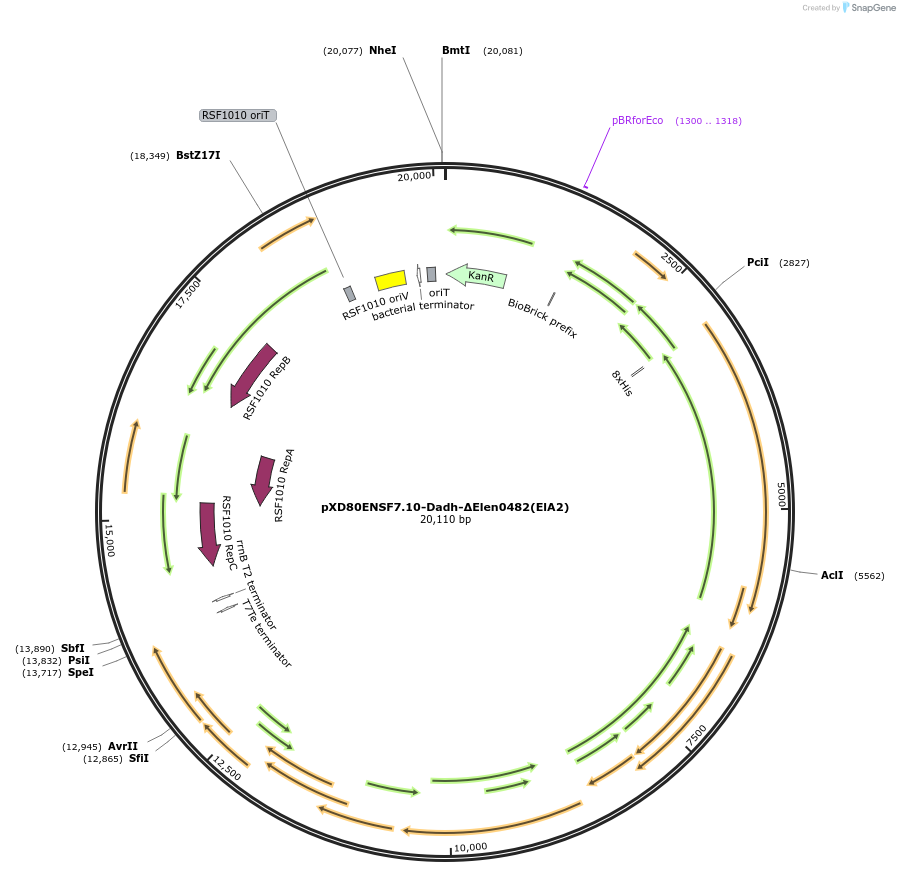
pXD80ENSF7.10-Dadh-ΔElen0482(ElA2)
Plasmid#225325PurposeExpresses Eggerthella lenta A2 Dadh (ΔElen_0482) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertDadh (ΔElen_0482)
TagsHis8ExpressionBacterialAvailable SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only