We narrowed to 13,148 results for: BASE
-
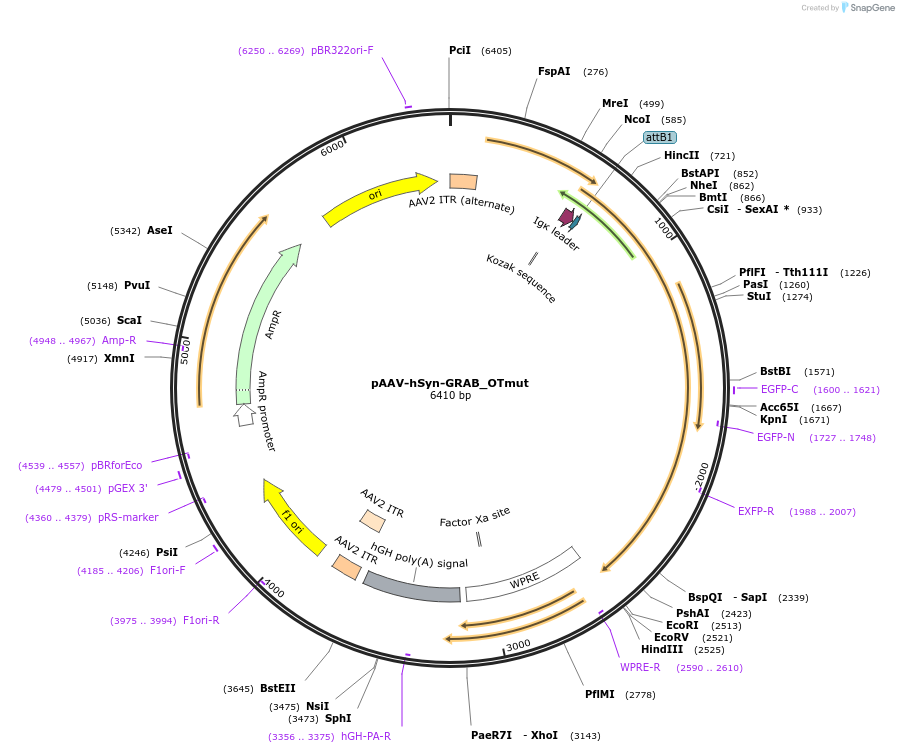
Plasmid#185388PurposeExpresses the genetically-encoded fluorescent oxytocin(OT) control sensor GRAB_OTmut in neuronsDepositorHas ServiceAAV1 and AAV9InsertGPCR activation based oxytocin control sensor GRAB_OTmut
UseAAVPromoterhSynAvailable SinceSept. 20, 2022AvailabilityAcademic Institutions and Nonprofits only -
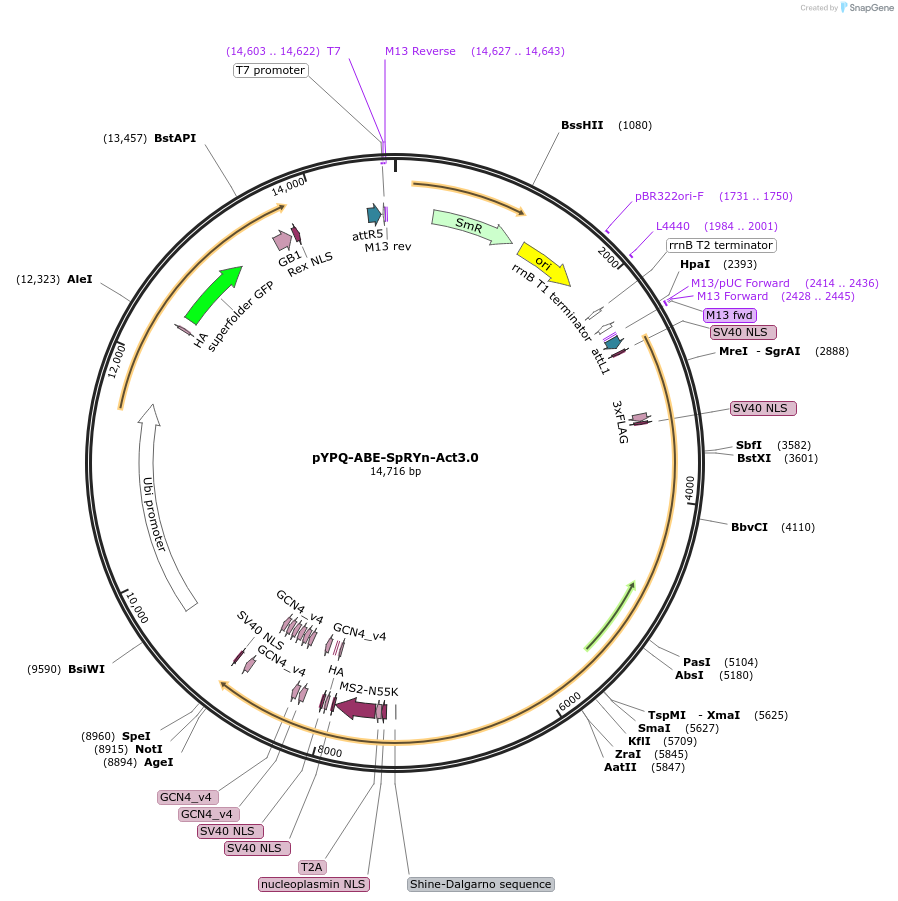
pYPQ-ABE-SpRYn-Act3.0
Plasmid#178959PurposeIt consists of a SpRY nickase (D10A) fused with an adenine deaminase (ABE8e) and MS2-SunTag-activators (ScFv-sfGFP-2xTAD), enabling simultaneous A to G conversion and gene activation.DepositorInsertABE8e-SpRY-T2A-MS2-SunTag-rbcS-E9t-ter-ZmUbi-scFv-sfGFP-2xTAD-GB1-NOS-ter
UseCRISPRExpressionPlantAvailable SinceMay 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
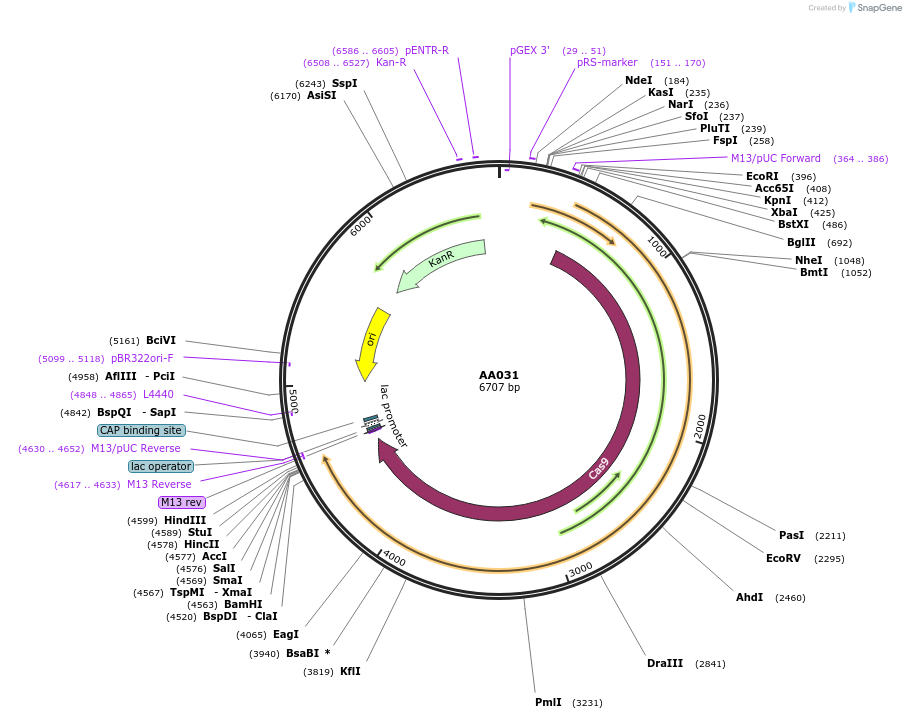
AA031
Plasmid#216011PurposeFragmid fragment: (Cas protein) nickase Cas for base editingDepositorHas ServiceCloning Grade DNAInsertnCas9-SpRY (D10A)_v1.1 [Sp]
UseCRISPR; FragmentAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
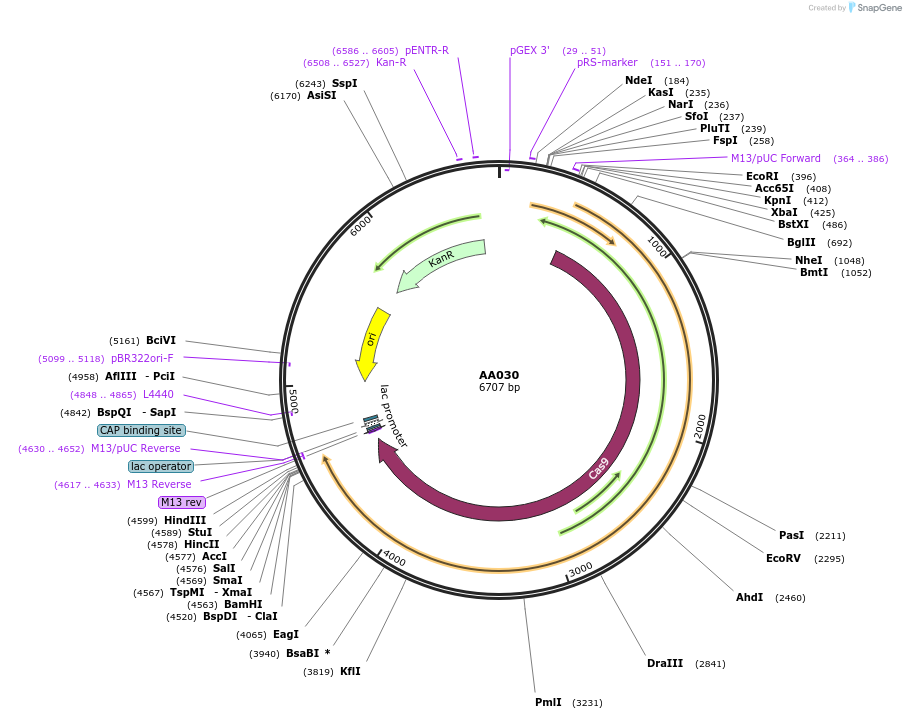
AA030
Plasmid#216010PurposeFragmid fragment: (Cas protein) nickase Cas for base editingDepositorHas ServiceCloning Grade DNAInsertnCas9-SpG (D10A)_v1.1 [Sp]
UseCRISPR; FragmentAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
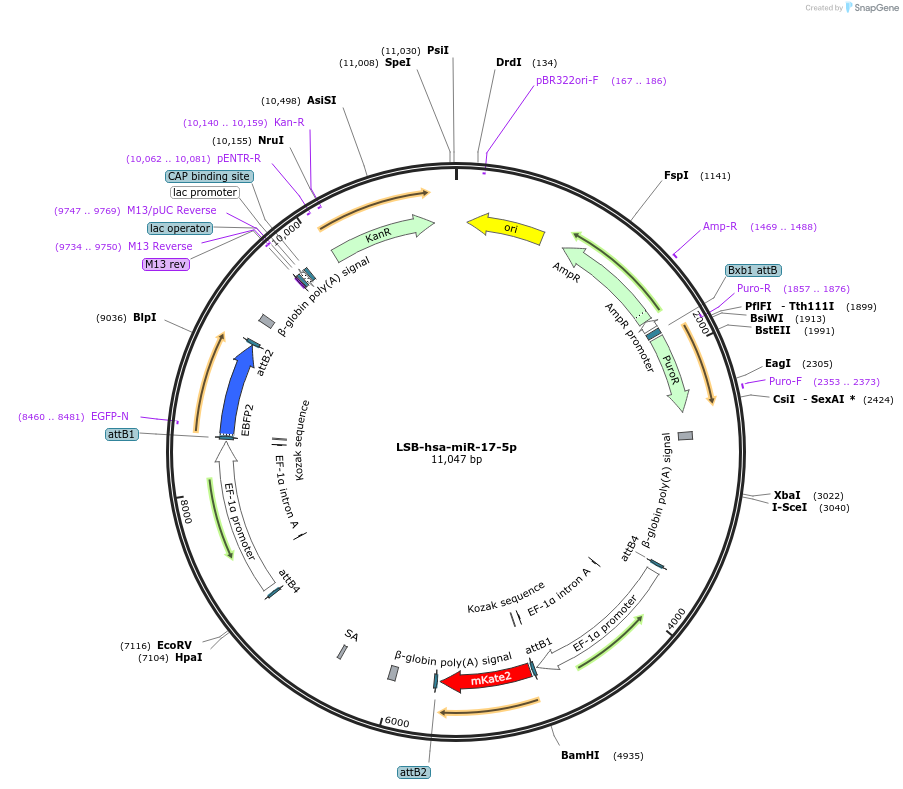
LSB-hsa-miR-17-5p
Plasmid#103274PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-17-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
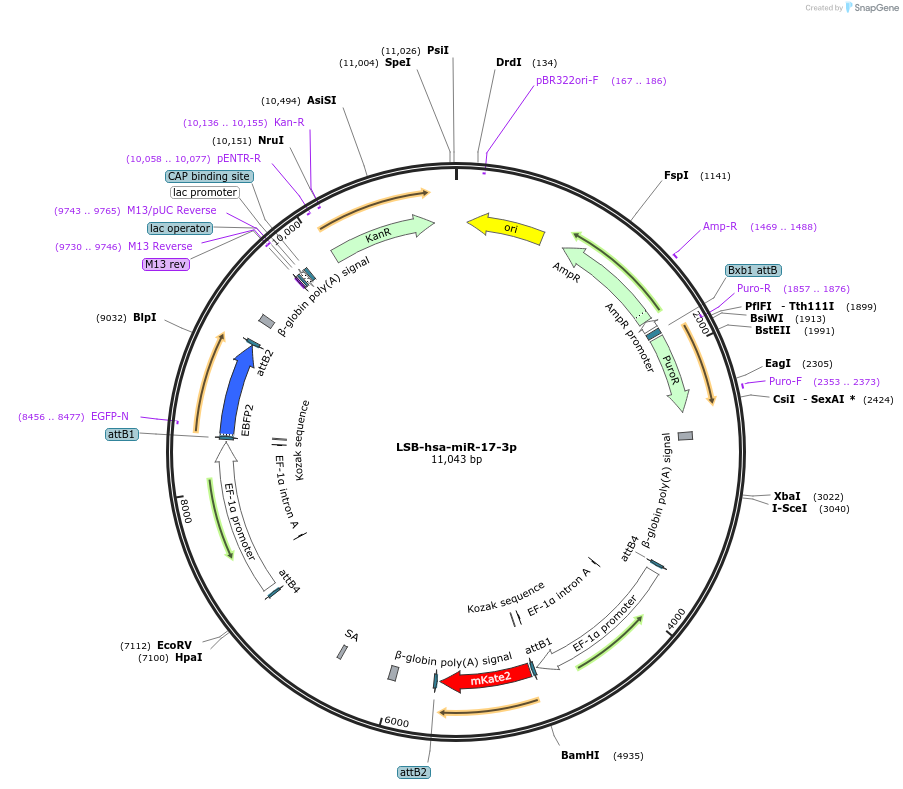
LSB-hsa-miR-17-3p
Plasmid#103273PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-17-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
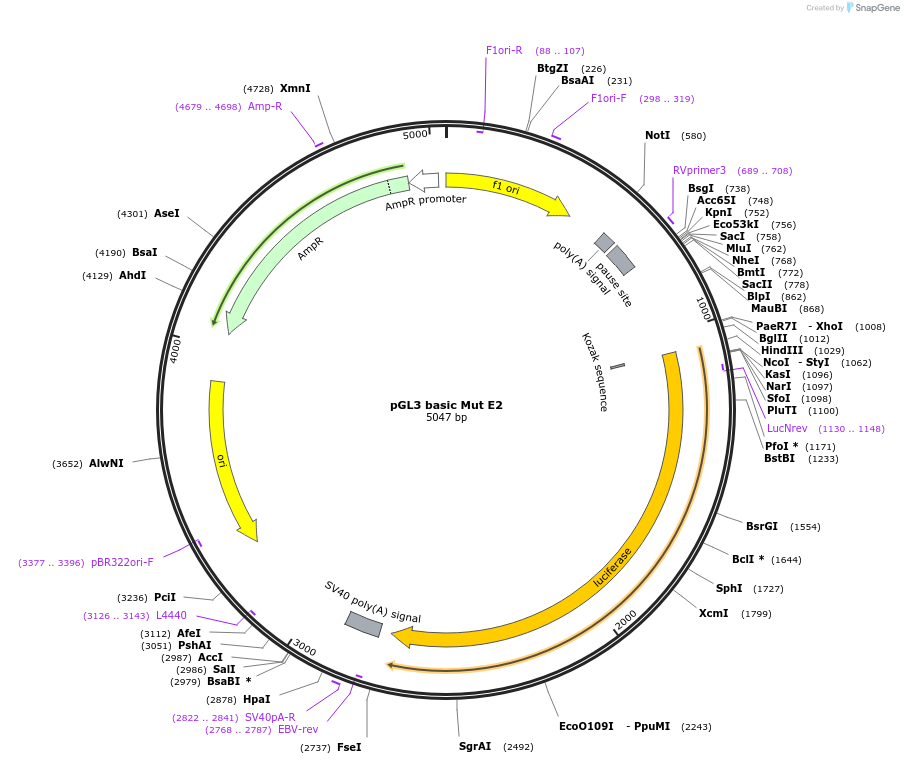
pGL3 basic Mut E2
Plasmid#48748PurposeExpression of luciferase driven by mPer2 promoter fragment containing mutated E-box2 (bases -112 to +98 with respect to transcription start site at +1)DepositorInsertmPer2 promoter/enhancer region containing mutated E-box2 (-112 to +98)
UseLuciferaseMutationMutated E-box2 sequence from CACGTT to GCTAGTAvailable SinceDec. 5, 2013AvailabilityAcademic Institutions and Nonprofits only -
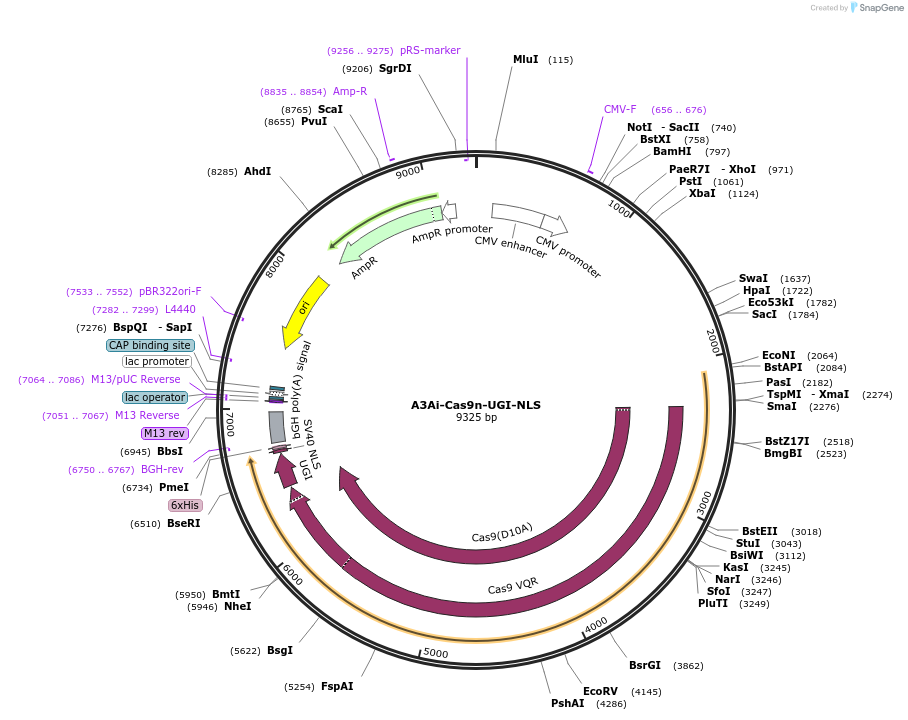
A3Ai-Cas9n-UGI-NLS
Plasmid#109425PurposeExpresses human APOBEC3A containing an L1 intron fused to Cas9n, Uracil DNA Glycosylase Inhibitor, with a nuclear localization signal.DepositorInsertApolipoprotein B mRNA Editing Enzyme, Catalytic Polypeptide-Like 3A (APOBEC3A Human)
UseCRISPRExpressionMammalianMutationInsertion of an L1 intron into the coding sequenc…PromoterCMVAvailable SinceJuly 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
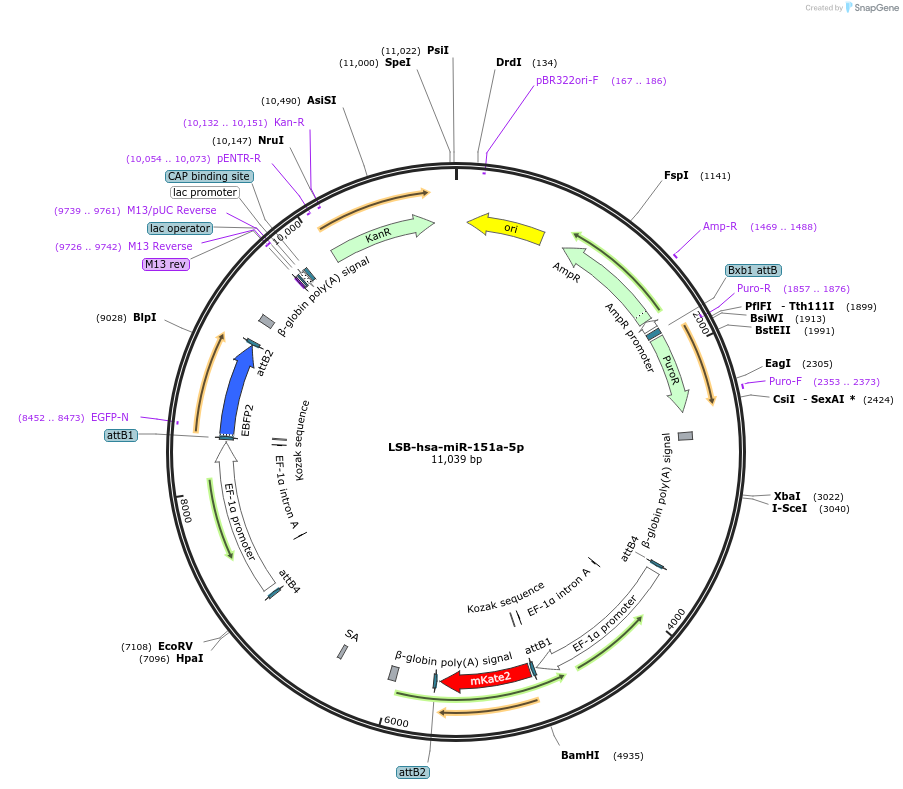
LSB-hsa-miR-151a-5p
Plasmid#103259PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-151a-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-151a-5p target (MIR151A Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
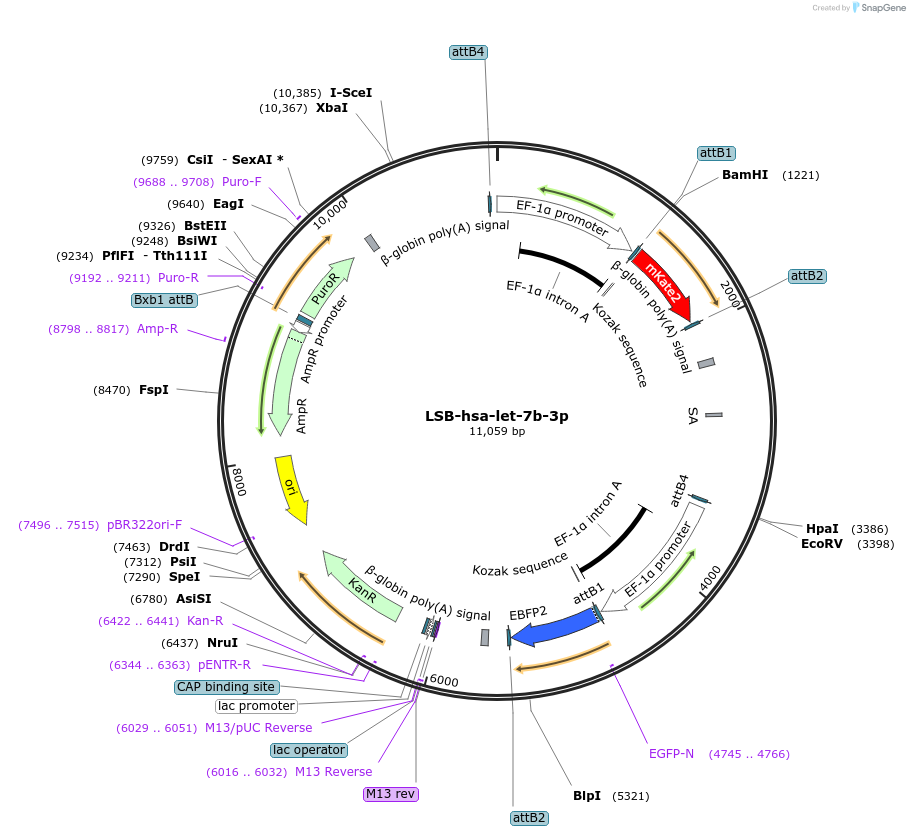
LSB-hsa-let-7b-3p
Plasmid#103147PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-let-7b-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-let-7b-3p target (MIRLET7B Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only