We narrowed to 27,024 results for: Cat
-
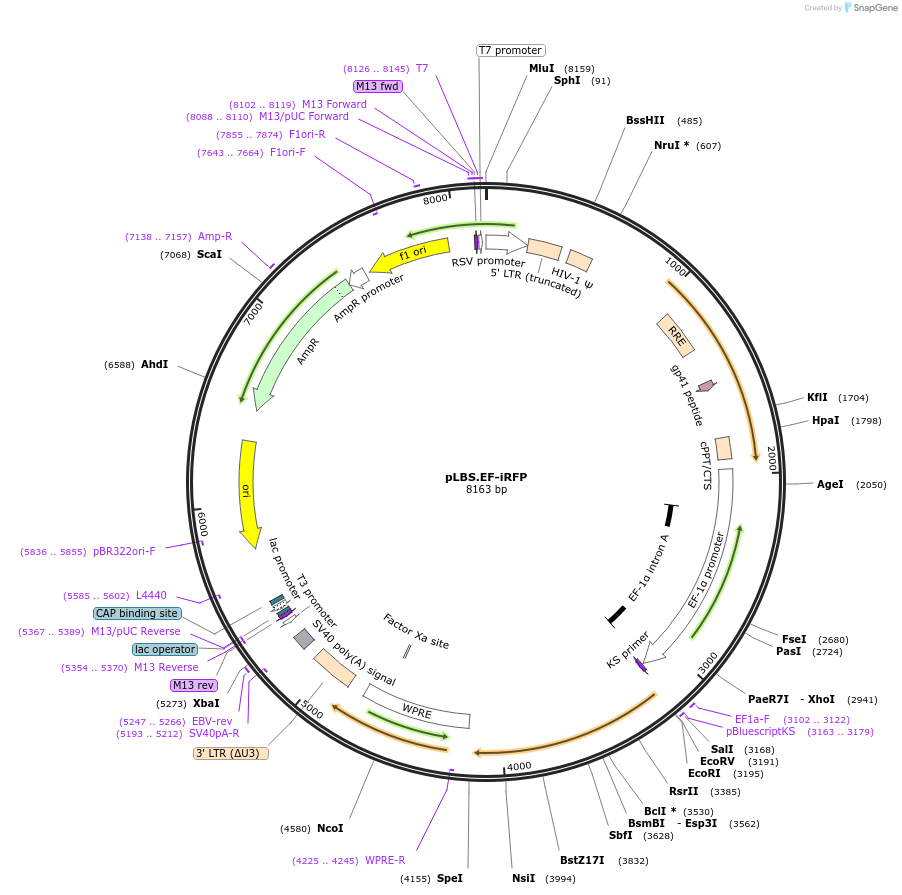
Plasmid#129329Purposecytosol marker, lentivirusDepositorInsertiRFP (from Dr. M. Matsuda)
UseLentiviralAvailable SinceOct. 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
mPA-GFP-tdTomato-FRET-10
Plasmid#58155PurposeLocalization: Cytosolic FRET, Excitation: 504 / 554, Emission: 517 / 581DepositorTypeEmpty backboneTagsmPA-GFP and tdTomatoExpressionMammalianAvailable SinceApril 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
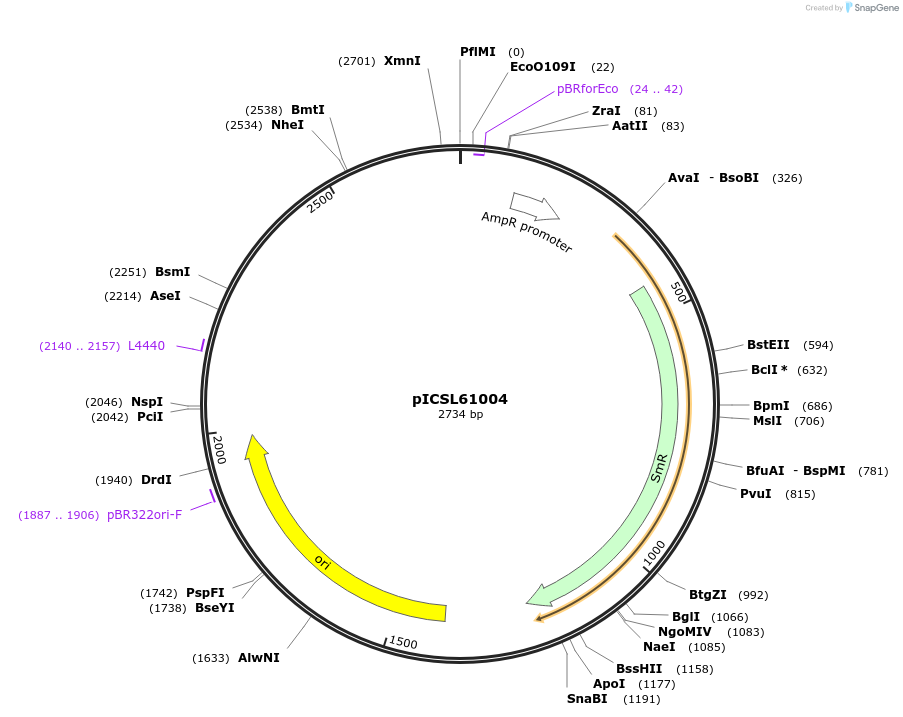
pICSL61004
Plasmid#245681PurposeLevel 0 Golden Gate part 3' Extensin-Truncated-EU (TerP1) terminator P1, GCTT - TAGADepositorInsert3' Extensin-Truncated-EU (TerP1) terminator P1
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
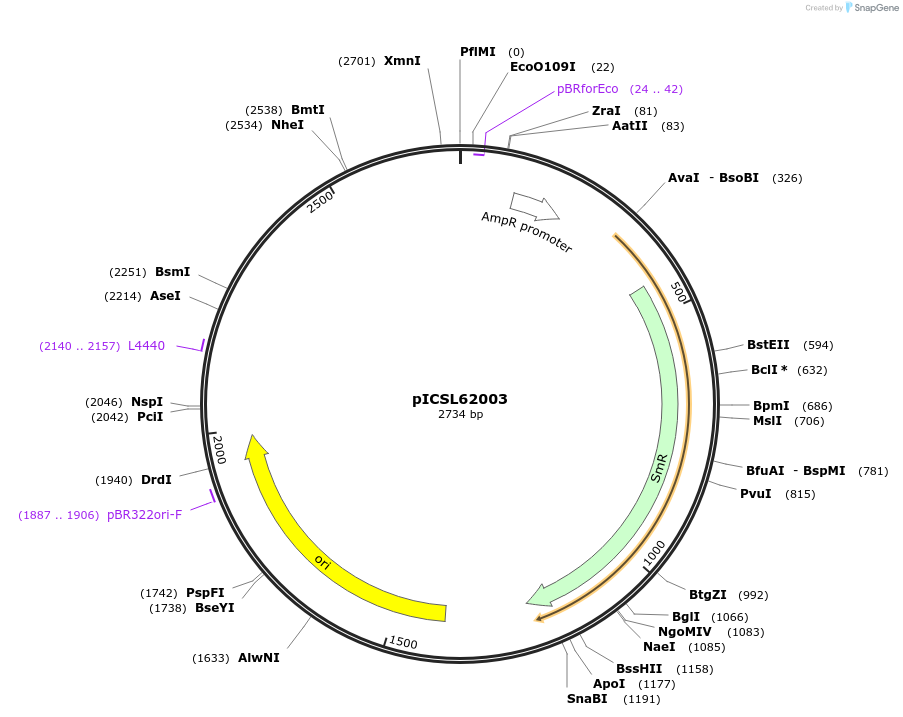
pICSL62003
Plasmid#245684PurposeLevel 0 Golden Gate part 3' Extensin-Truncated (TerP2) terminator P2, TAGA - CGCTDepositorInsert3' Extensin-Truncated (TerP2) terminator P2
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
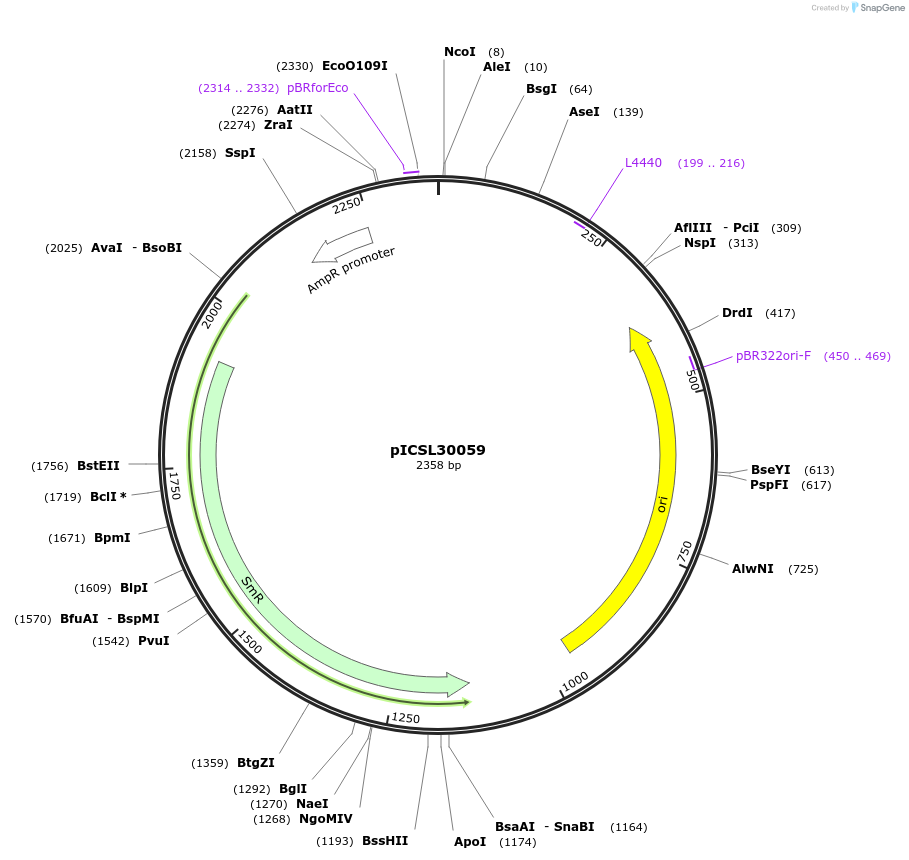
pICSL30059
Plasmid#245562PurposeLevel 0 Golden Gate part N-Terminal Tag with overhangs CCAT - AATGDepositorInsertN-Terminal tag Self-Cleaving tag compatible with ProteinSelect purification system
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
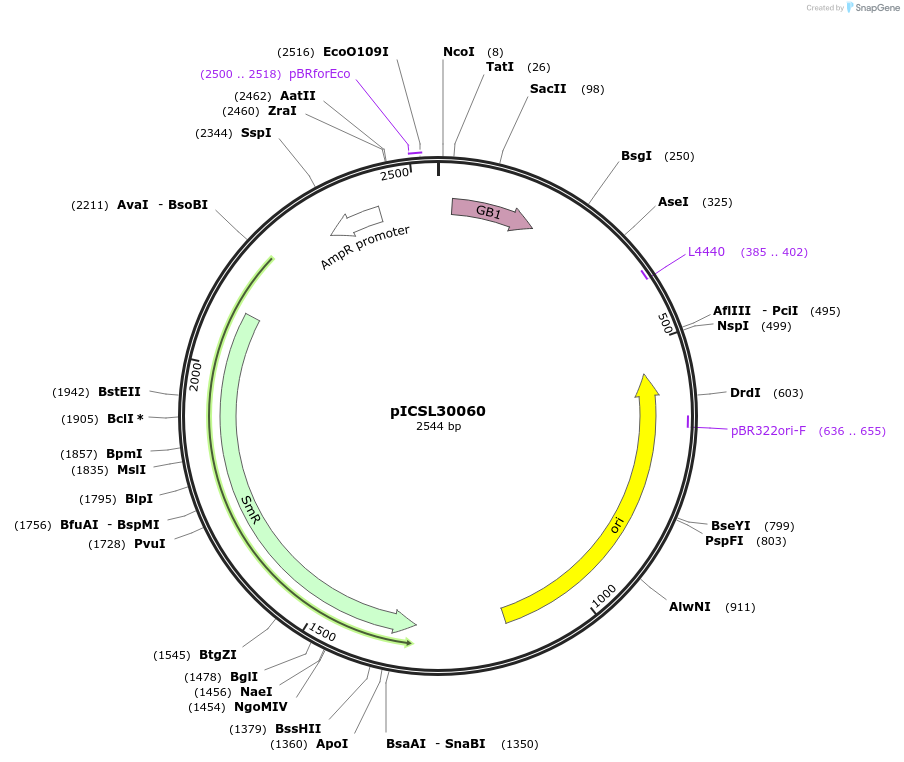
pICSL30060
Plasmid#245563PurposeLevel 0 Golden Gate part N-Terminal Tag with overhangs CCAT - AATGDepositorInsertN-Terminal tag Self-Cleaving GB1 tag compatible with ProteinSelect purification system
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
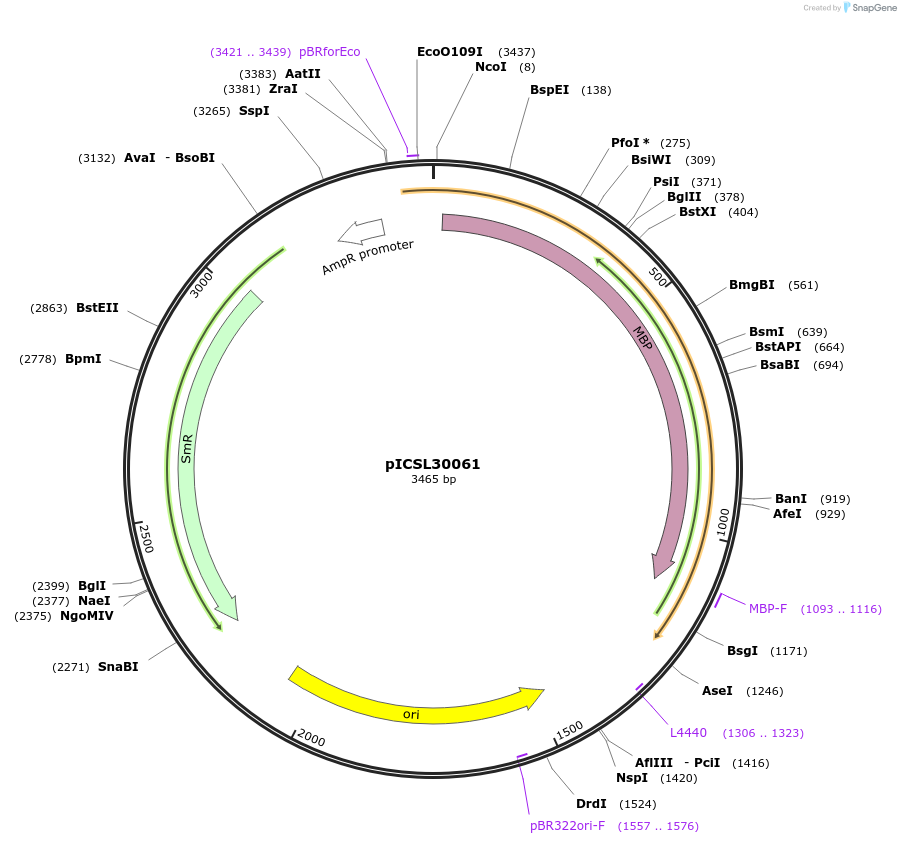
pICSL30061
Plasmid#245564PurposeLevel 0 Golden Gate part N-Terminal Tag with overhangs CCAT - AATGDepositorInsertN-Terminal tag Self-Cleaving MBP tag compatible with ProteinSelect purification system
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
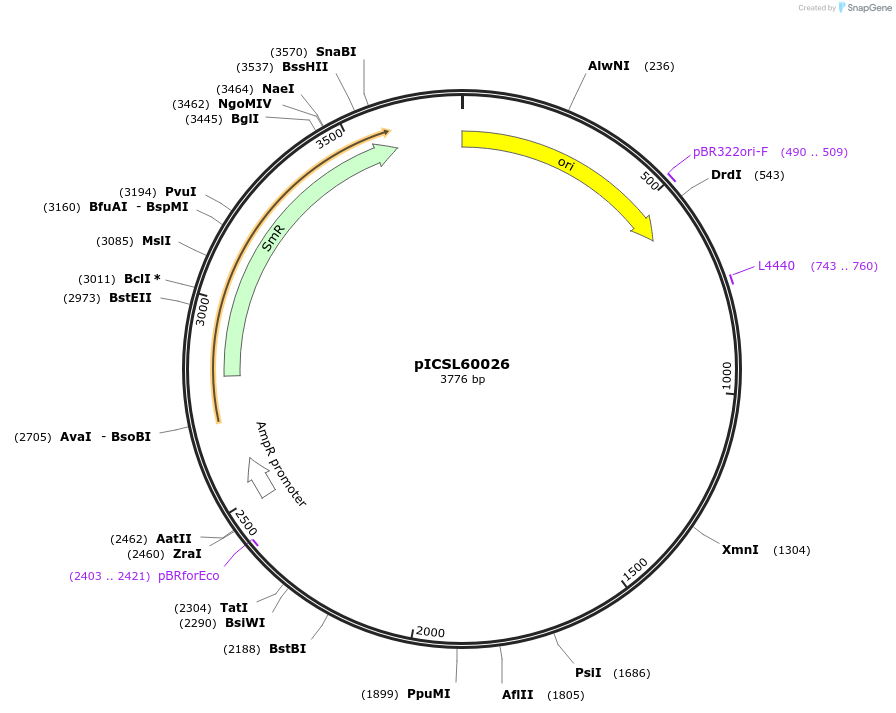
pICSL60026
Plasmid#245674PurposeLevel 0 Golden Gate part Extensin-Truncated-EU + NbACT terminator, GCTT - CGCTDepositorInsertExtensin-Truncated-EU + NbACT terminator
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
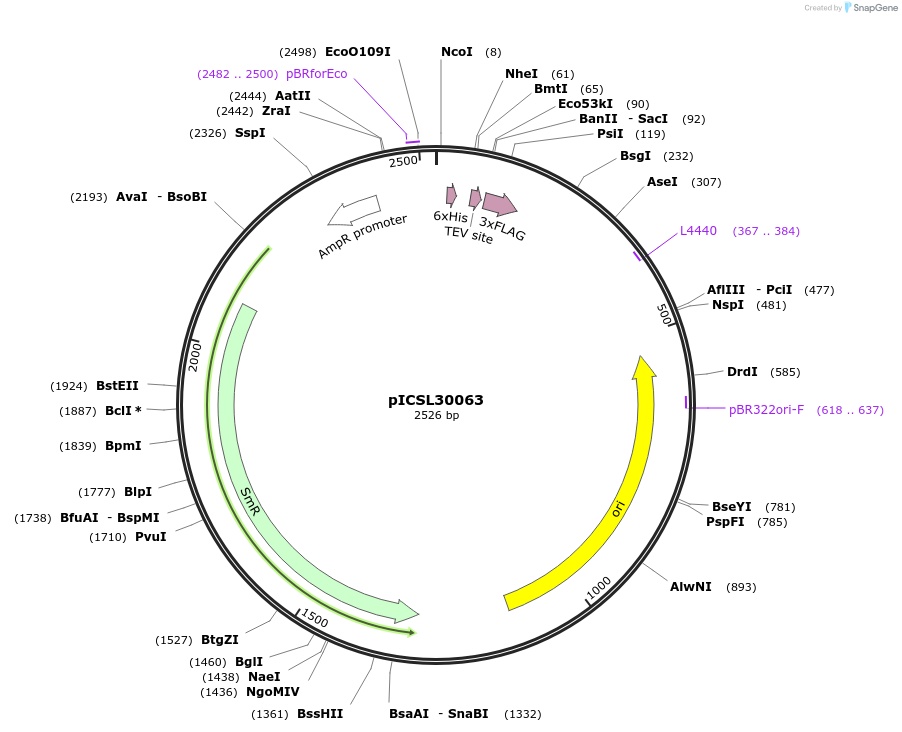
pICSL30063
Plasmid#245566PurposeLevel 0 Golden Gate part N-Terminal Tag with overhangs CCAT - AATGDepositorInsertN-Terminal tag Self-Cleaving HF tag compatible with ProteinSelect purification system
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
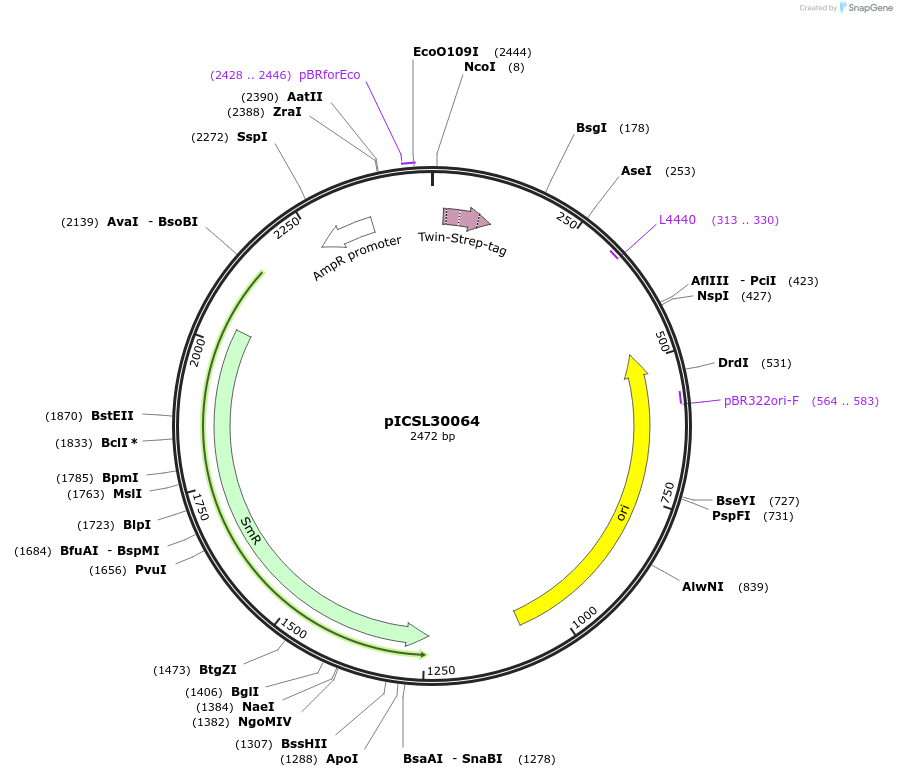
pICSL30064
Plasmid#245567PurposeLevel 0 Golden Gate part N-Terminal Tag with overhangs CCAT - AATGDepositorInsertN-Terminal tag Self-Cleaving STREPII tag compatible with ProteinSelect purification system
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
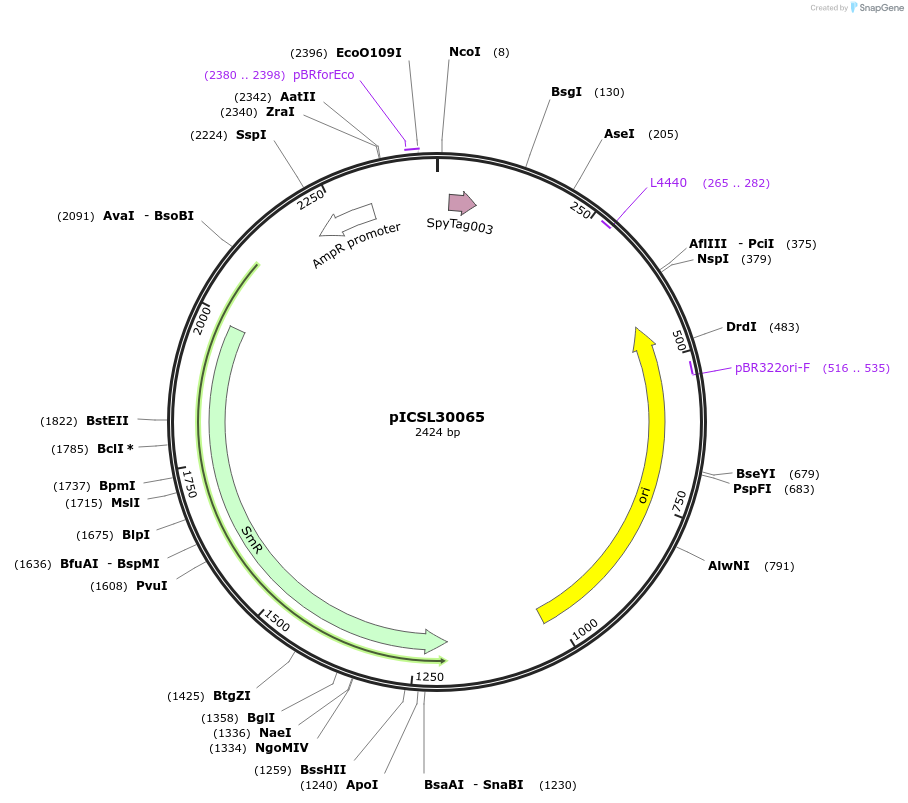
pICSL30065
Plasmid#245568PurposeLevel 0 Golden Gate part N-Terminal Tag with overhangs CCAT - AATGDepositorInsertN-Terminal tag Self-Cleaving SPY tag compatible with ProteinSelect purification system
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
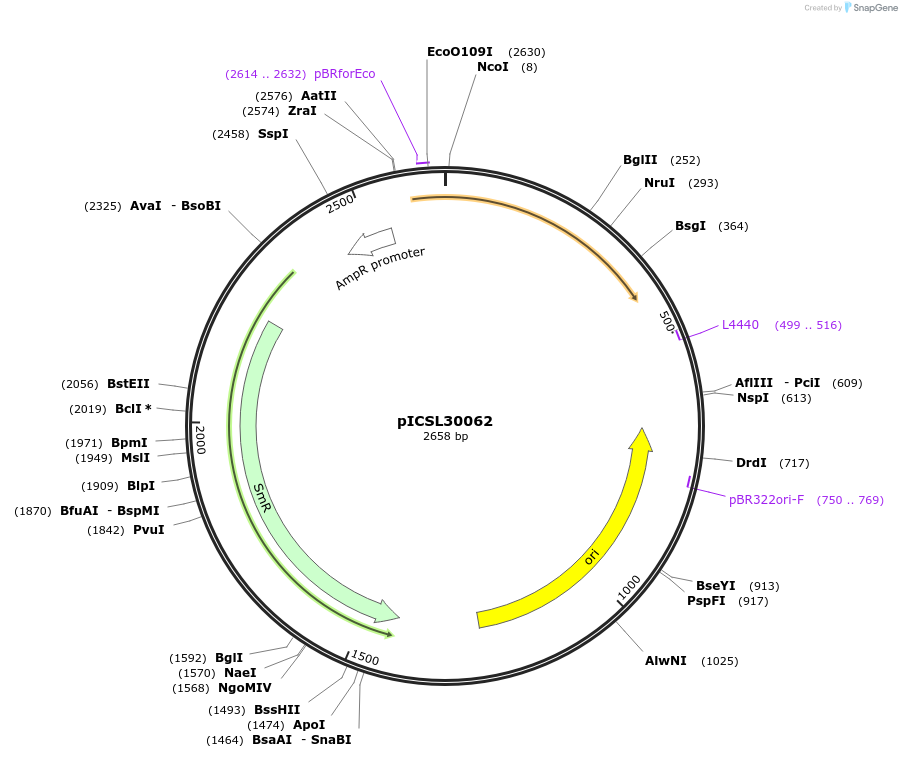
pICSL30062
Plasmid#245565PurposeLevel 0 Golden Gate part N-Terminal Tag with overhangs CCAT - AATGDepositorInsertN-Terminal tag Self-Cleaving SUMO tag compatible with ProteinSelect purification system
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
1203T-CMV-Cas7.1-NLS-bGHpolyA
Plasmid#221540PurposeEffector only expresses Cas7.1 domain (truncated from DiCas7-11 effector), nuclear localization signal sequence on 5' and 3' end of the coding domainDepositorInsertTruncated DiCas7-11
UseCRISPRExpressionMammalianAvailable SinceFeb. 19, 2025AvailabilityAcademic Institutions and Nonprofits only -
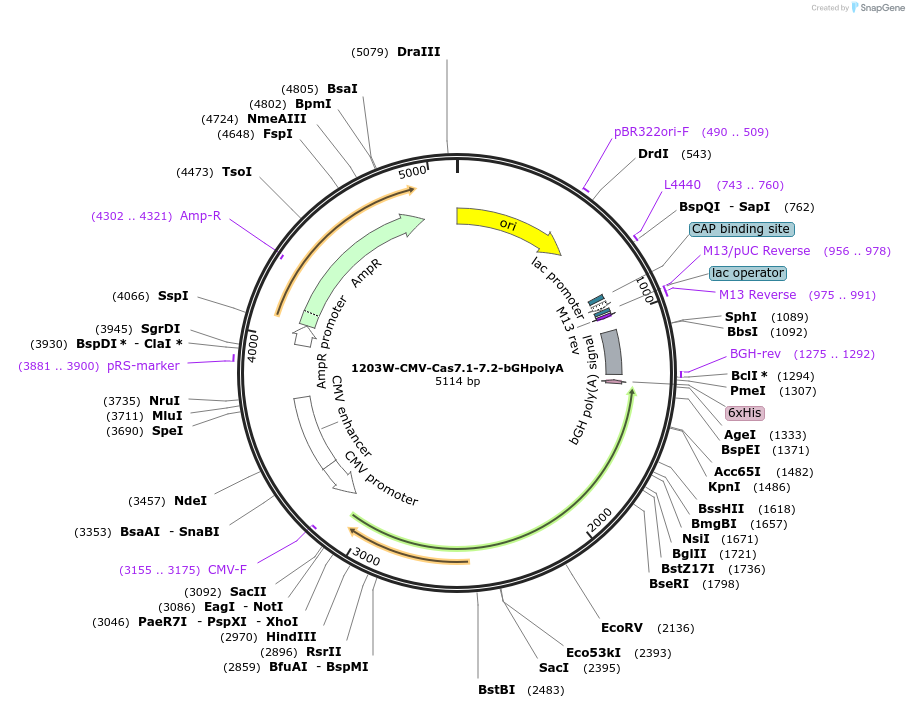
1203W-CMV-Cas7.1-7.2-bGHpolyA
Plasmid#220961PurposeEffector expresses Cas7.1, Linker1, Cas11, Linker 2, and Cas7.2 domains from DiCas7-11, deactivated (truncated from DiCas7-11 effector)DepositorInsertTruncated DiCas7-11
UseCRISPRExpressionMammalianAvailable SinceFeb. 19, 2025AvailabilityAcademic Institutions and Nonprofits only -
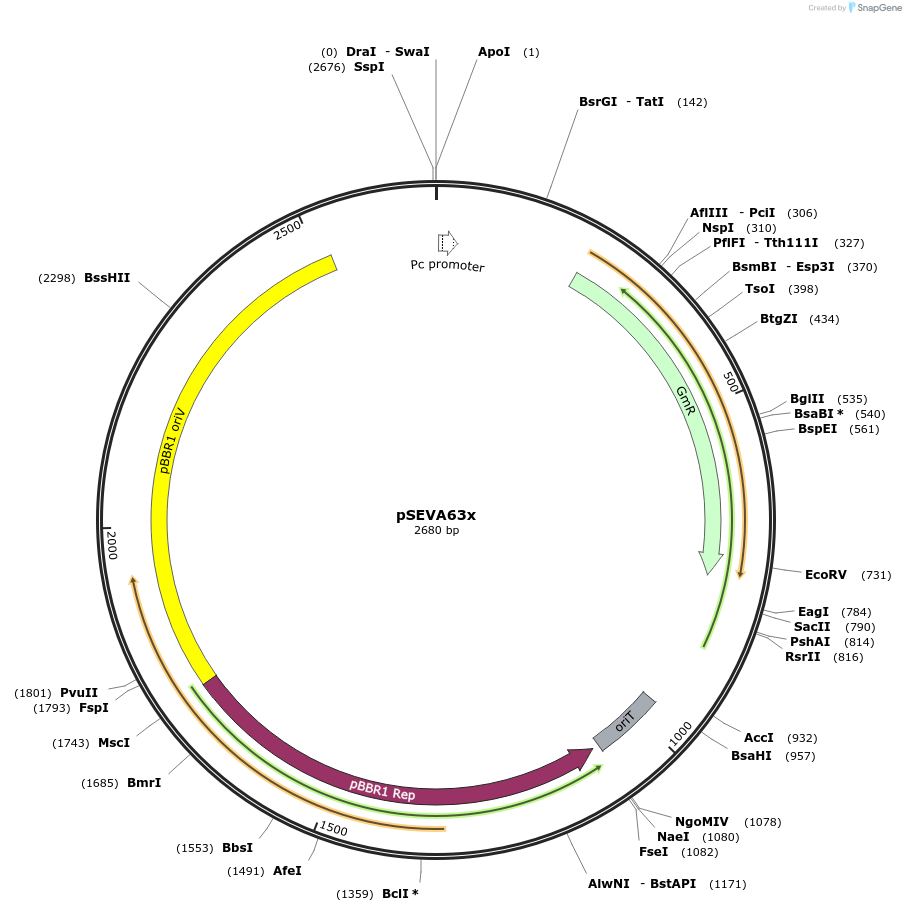
pSEVA63x
Plasmid#190437PurposeEmpty pSEVA63x vector.DepositorArticleTypeEmpty backboneUseSynthetic BiologyAvailable SinceDec. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
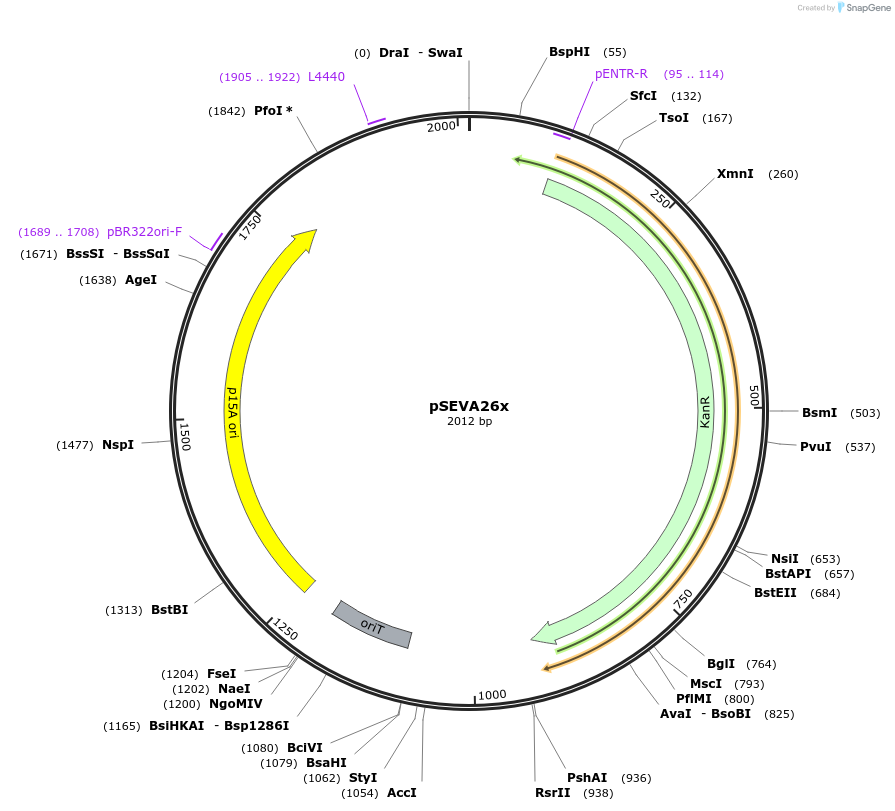
pSEVA26x
Plasmid#190438PurposeEmpty pSEVA26x vector.DepositorArticleTypeEmpty backboneUseSynthetic BiologyAvailable SinceDec. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
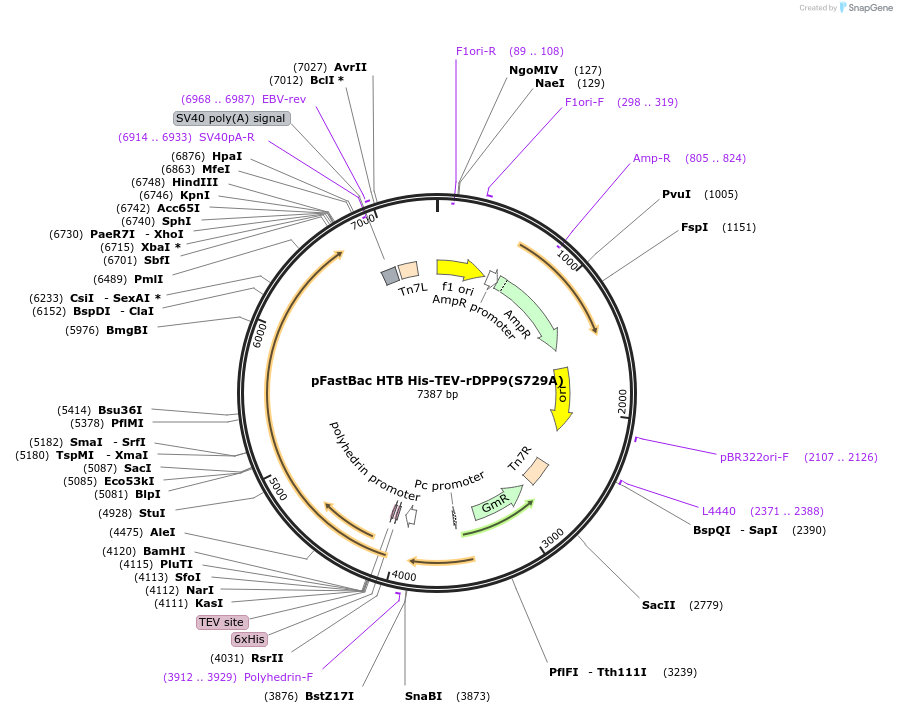
pFastBac HTB His-TEV-rDPP9(S729A)
Plasmid#217327PurposePurification of catalytically-dead rat DPP9 from insect cellsDepositorInsertDPP9
TagsHis-TEVExpressionInsectMutationS729APromoterpolyhedrinAvailable SinceAug. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
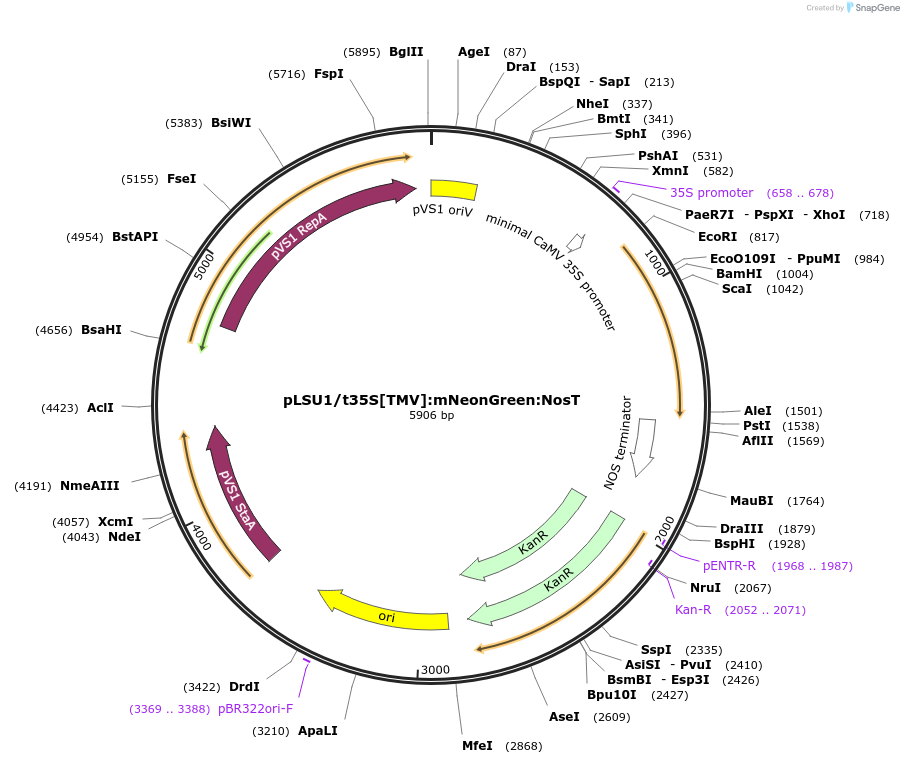
pLSU1/t35S[TMV]:mNeonGreen:NosT
Plasmid#212162PurposeThis binary vector expresses mNeonGreen with a truncated 35sCAMV promoter with strong UTR (Tomato Mosaic Virus)DepositorInsertTomato codon optimized version of mNeonGreen
ExpressionPlantMutationmNeonGreen codon-optimized for tomatoPromotertruncated 35sCaMVAvailable SinceMarch 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
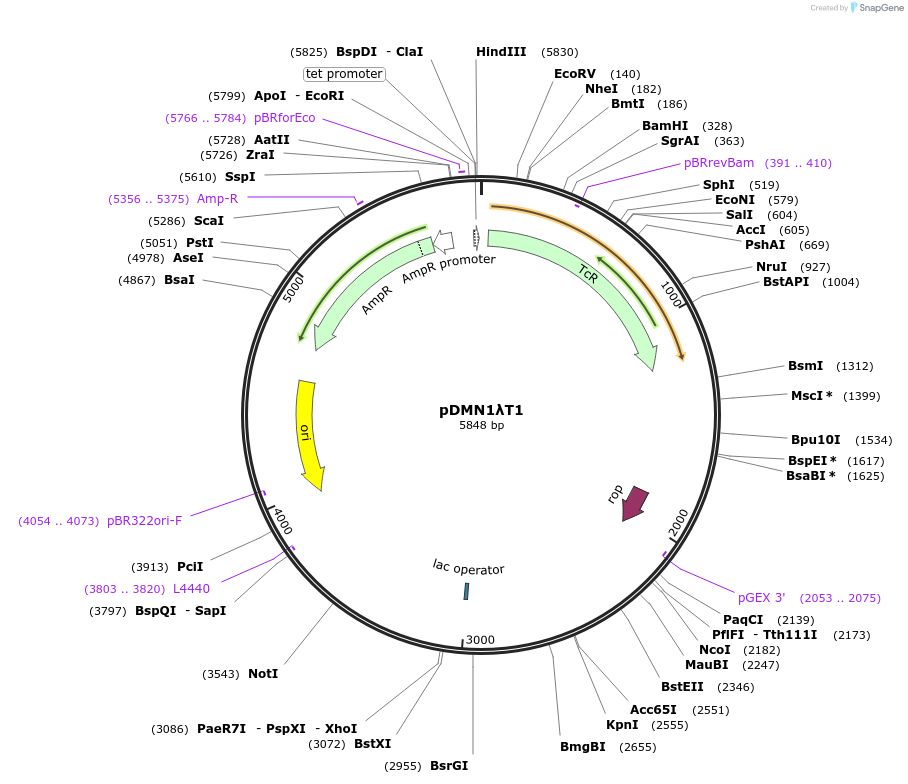
pDMN1λT1
Plasmid#211943Purposein vitro transcriptionDepositorInsertT7A1-O1-λT1
UseUnspecifiedPromoterT7A1Available SinceFeb. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
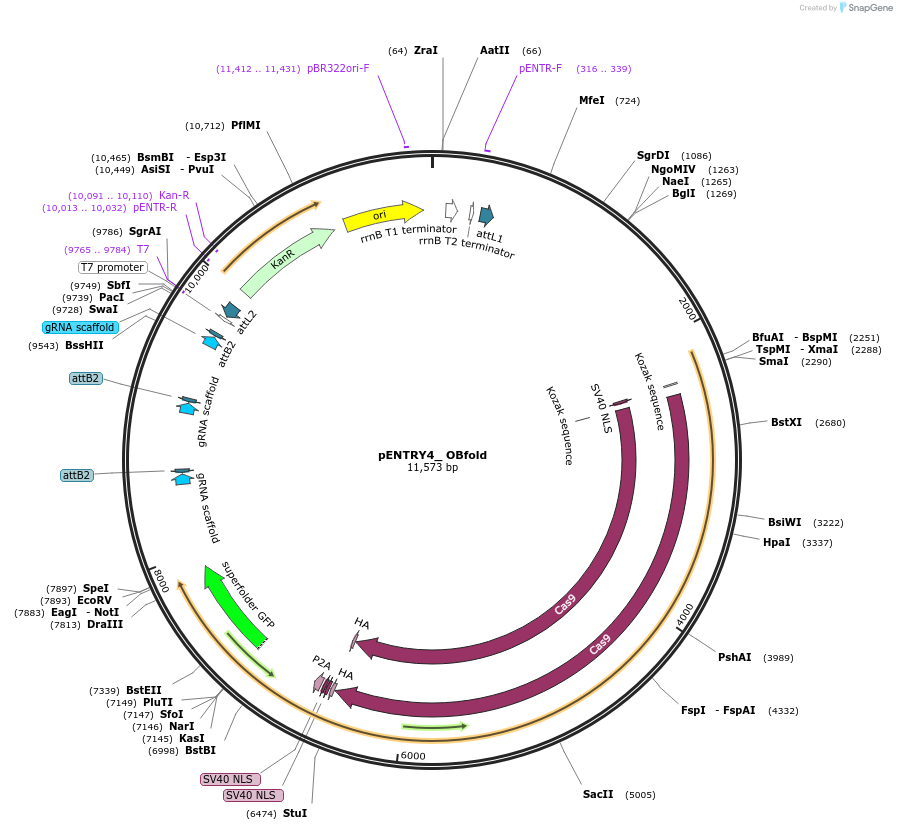
pENTRY4_ OBfold
Plasmid#126901PurposeGateway pENTR-plasmid to generate a binary construct for genome editing of the wheat homolog of a susceptibility gene of O. sativa. Encodes wheat optimized Cas9 and gene specific sgRNA modules.DepositorInsertWheat_live_Cas9
UseCRISPRAvailable SinceJuly 12, 2019AvailabilityAcademic Institutions and Nonprofits only