We narrowed to 27,048 results for: STI
-
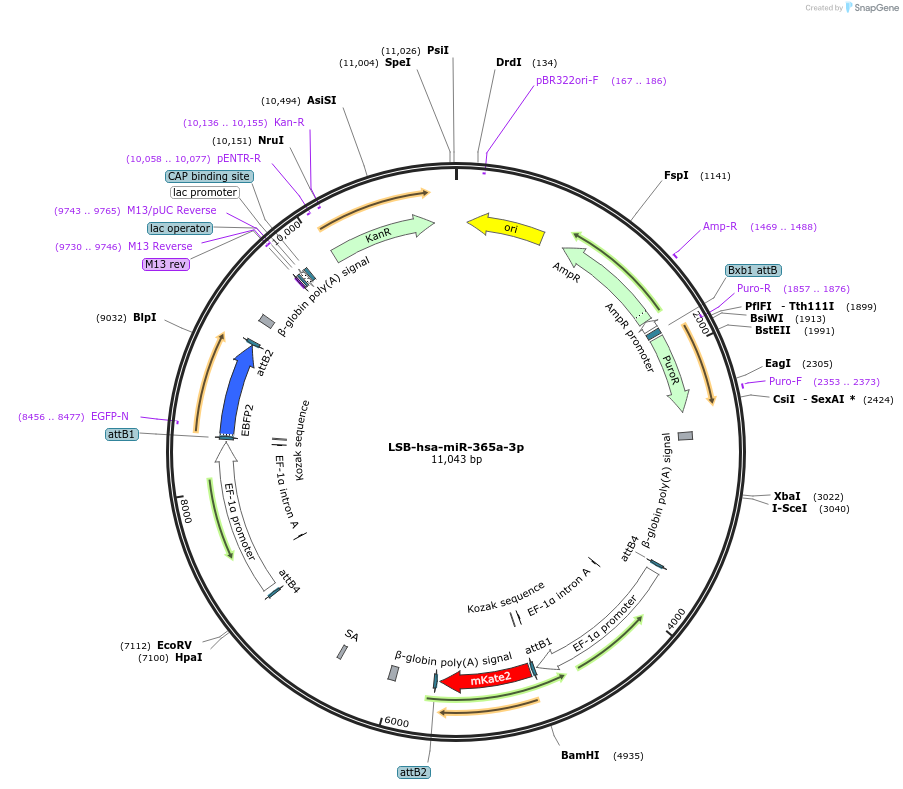
Plasmid#103482PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-365a-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-365a-3p target (MIR363 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
LSB-hsa-miR-369-5p
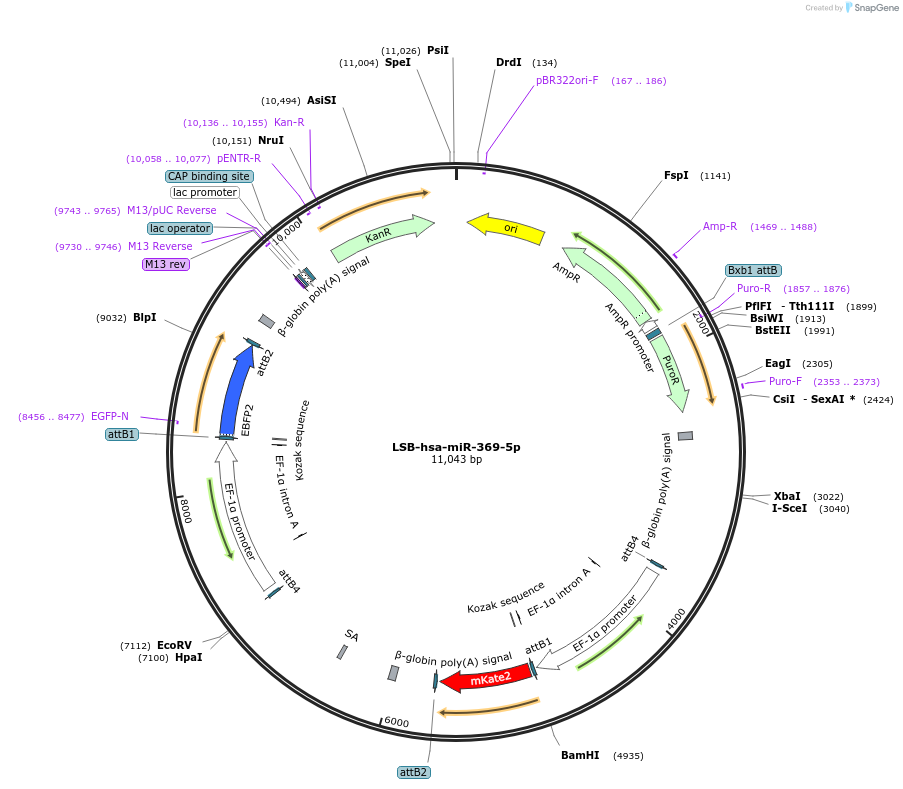
Plasmid#103487PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-369-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-369-5p target (MIR369 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
LSB-hsa-miR-370-3p
Plasmid#103488PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-370-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-370-3p target (MIR369 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
LSB-hsa-miR-374a-3p
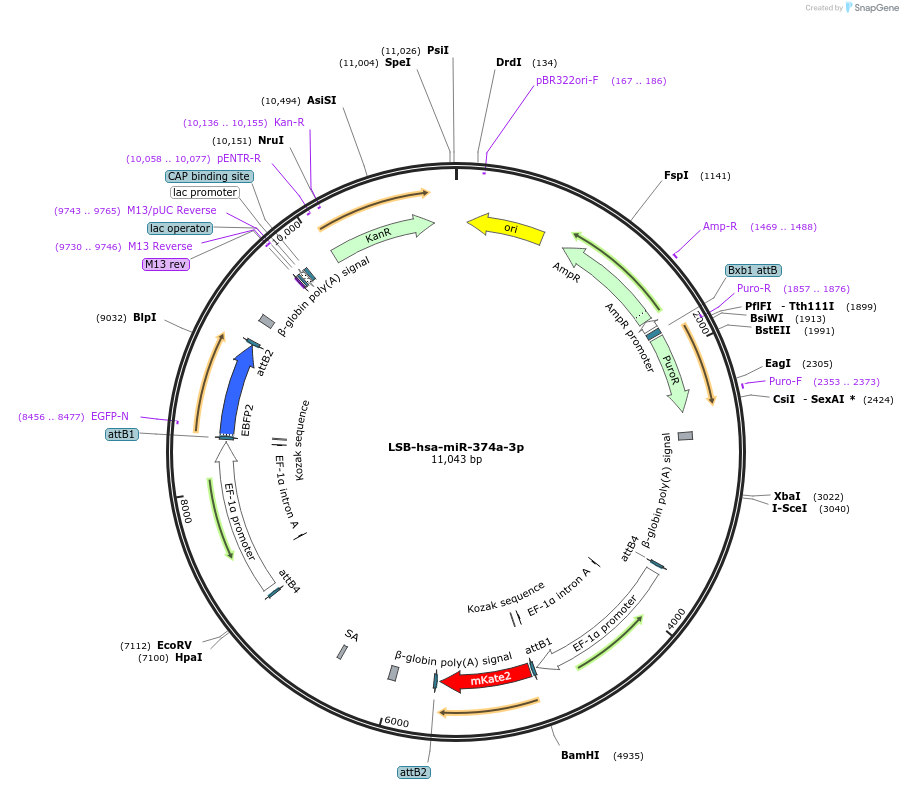
Plasmid#103490PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-374a-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-374a-3p target (MIR370 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
LSB-hsa-miR-374a-5p
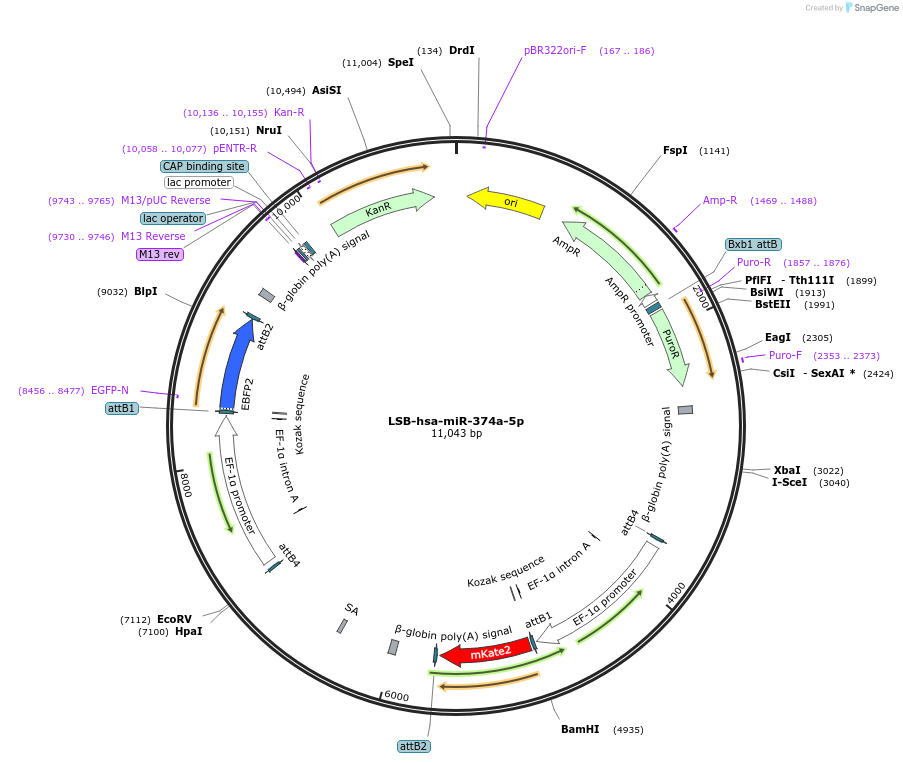
Plasmid#103491PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-374a-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-374a-5p target (MIR374A Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
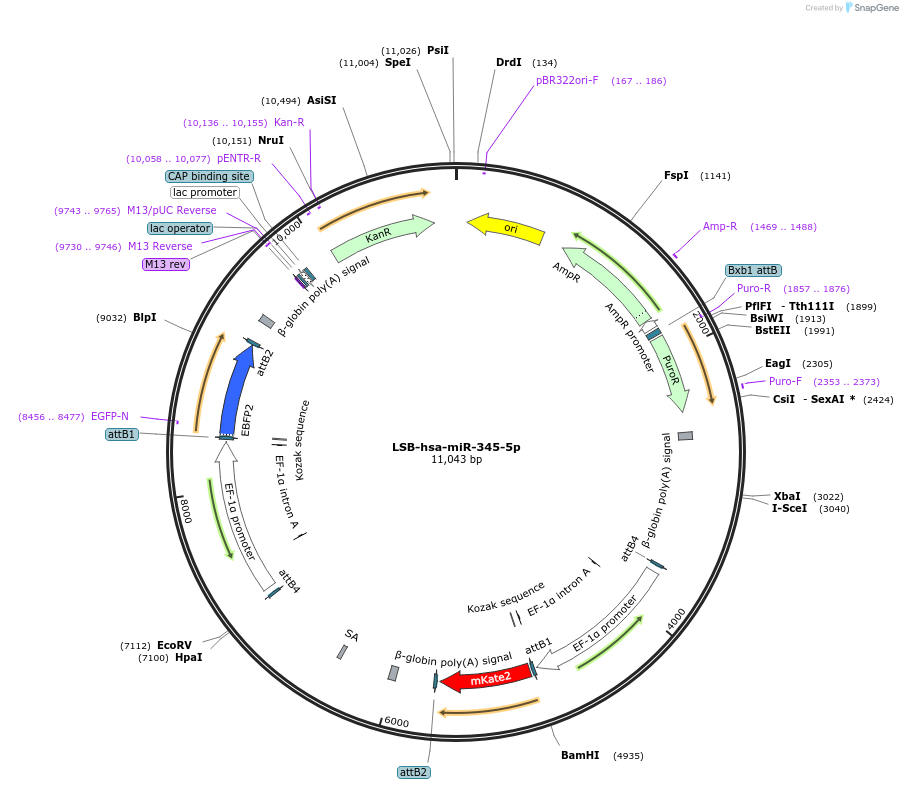
LSB-hsa-miR-345-5p
Plasmid#103463PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-345-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-345-5p target (MIR345 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
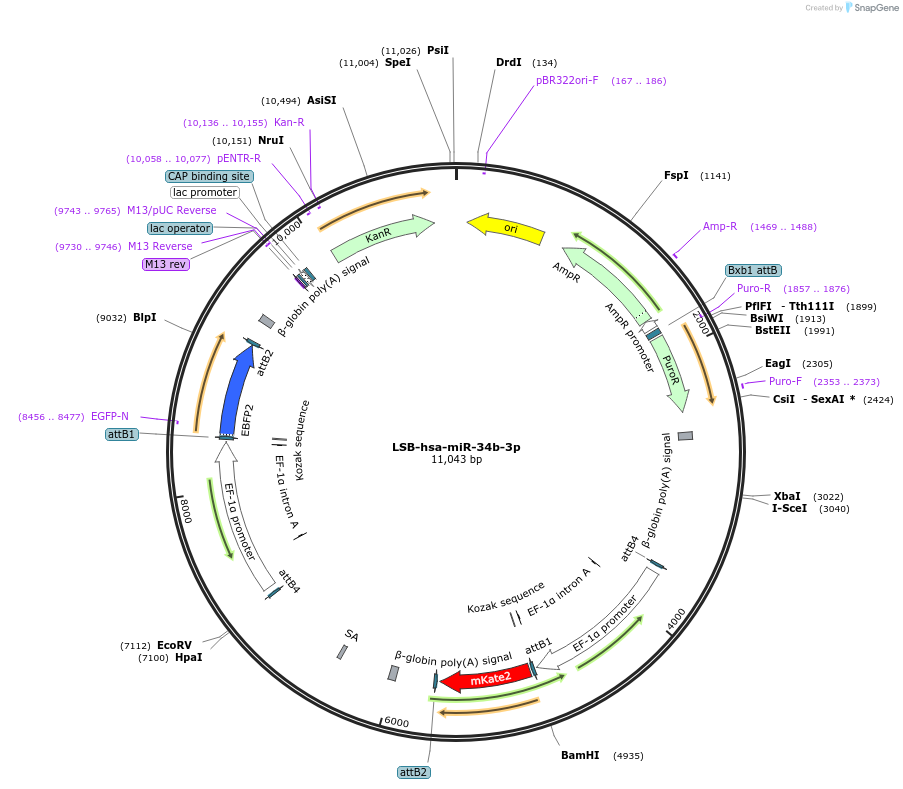
LSB-hsa-miR-34b-3p
Plasmid#103466PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-34b-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-34b-3p target (MIR34A Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
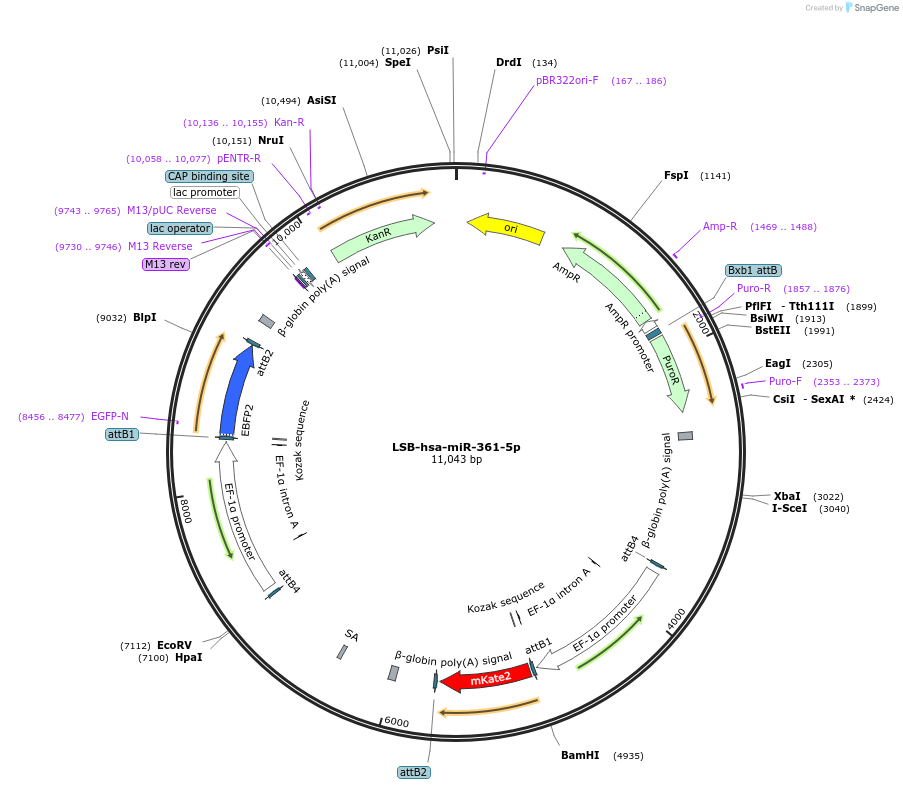
LSB-hsa-miR-361-5p
Plasmid#103473PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-361-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-361-5p target (MIR361 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
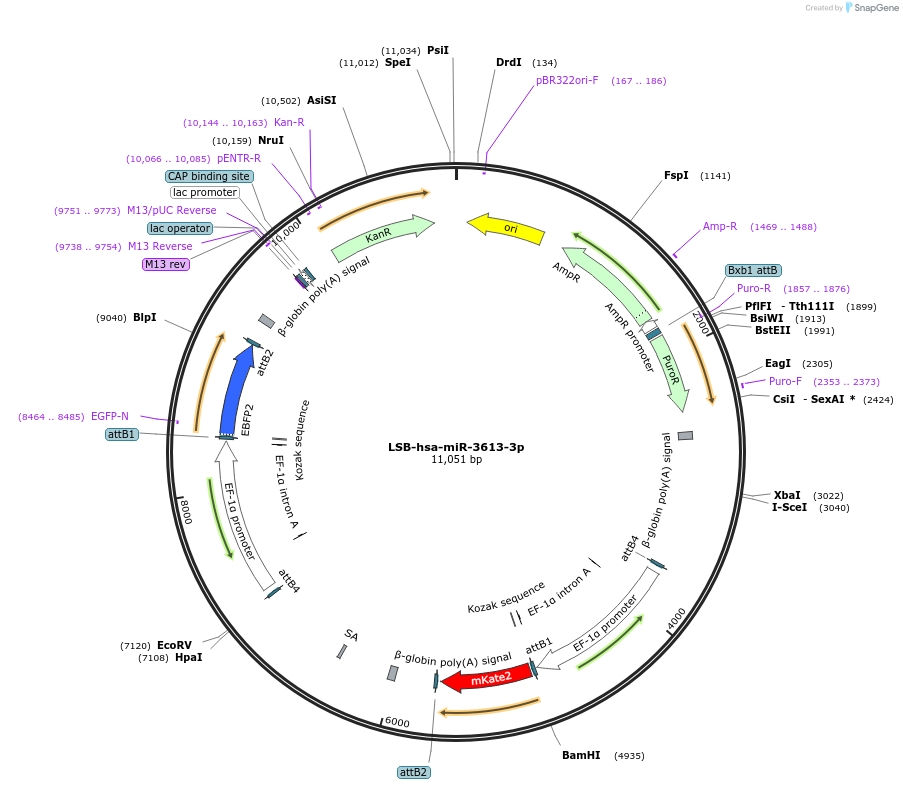
LSB-hsa-miR-3613-3p
Plasmid#103474PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-3613-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-3613-3p target (MIR361 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
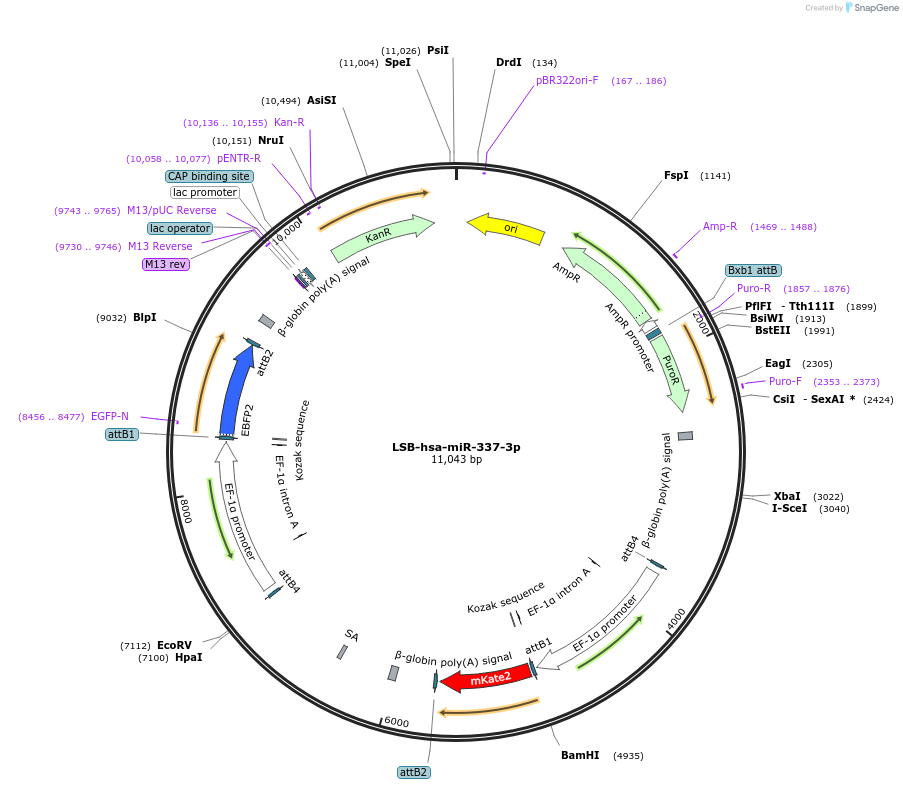
LSB-hsa-miR-337-3p
Plasmid#103448PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-337-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-337-3p target (MIR335 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
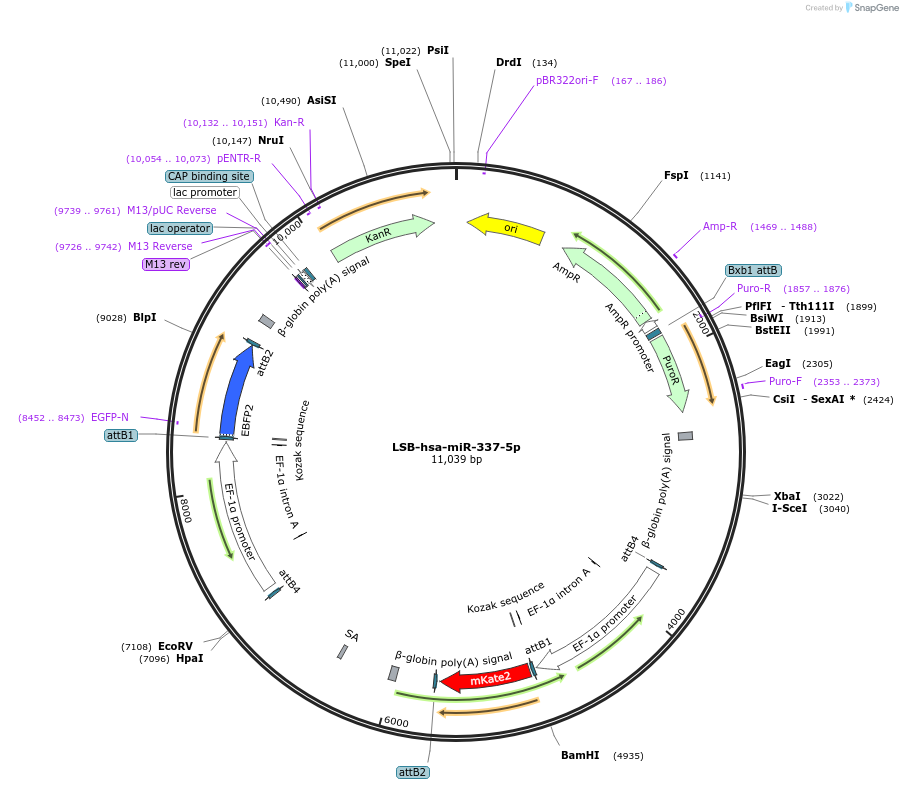
LSB-hsa-miR-337-5p
Plasmid#103449PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-337-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-337-5p target (MIR337 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
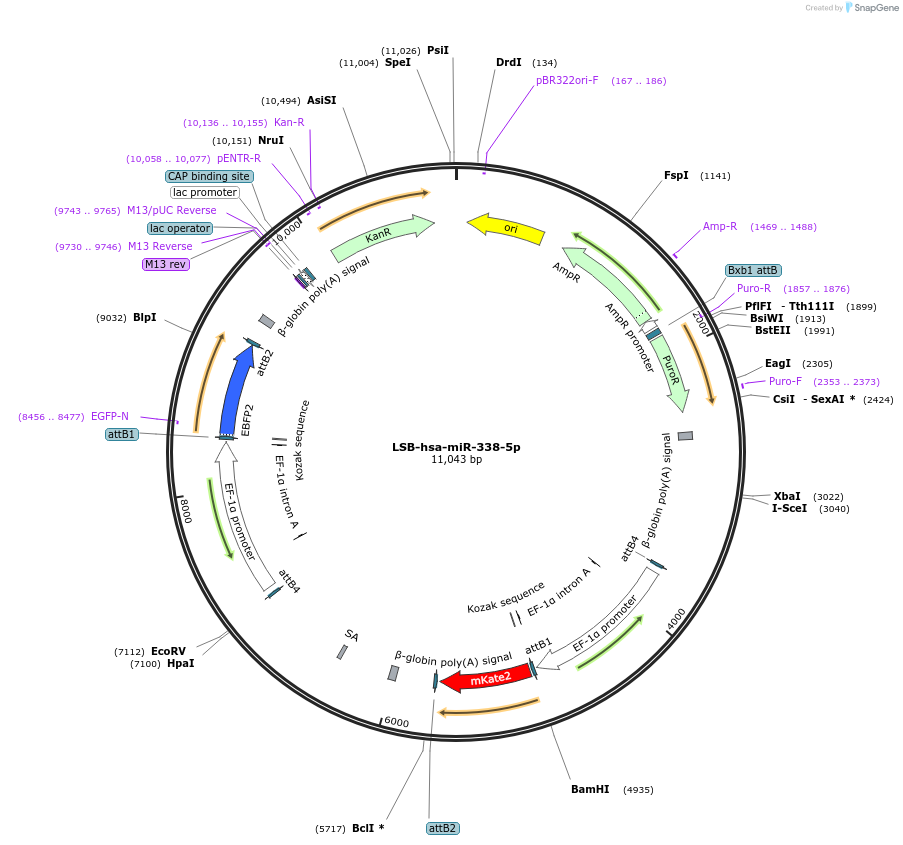
LSB-hsa-miR-338-5p
Plasmid#103451PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-338-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-338-5p target (MIR338 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
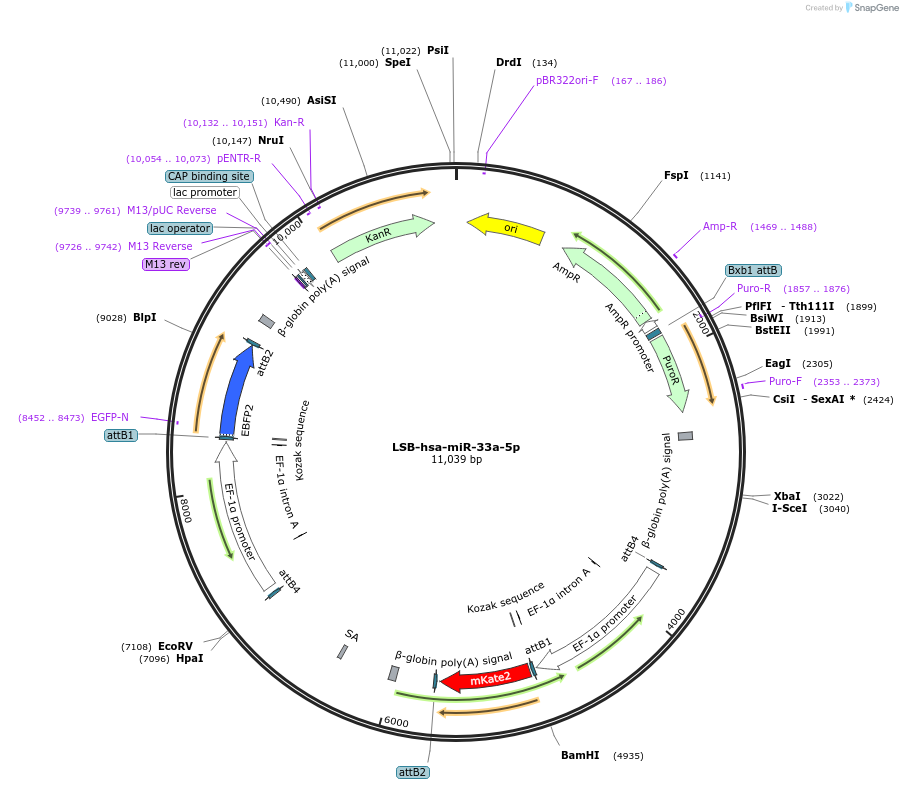
LSB-hsa-miR-33a-5p
Plasmid#103455PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-33a-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-33a-5p target (MIR33A Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
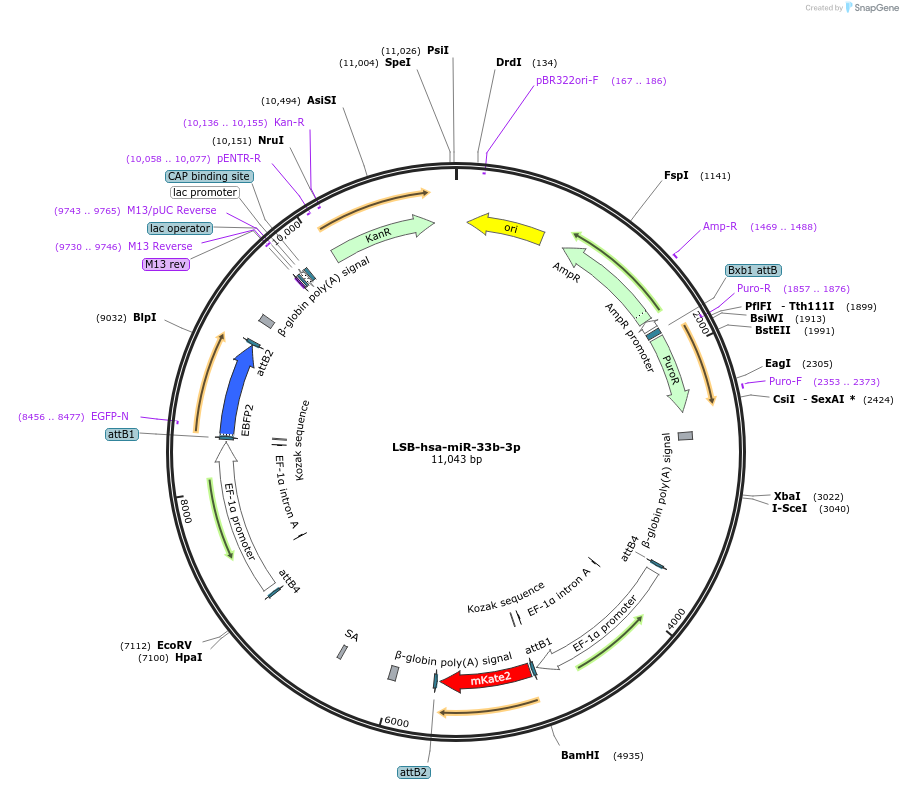
LSB-hsa-miR-33b-3p
Plasmid#103456PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-33b-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-33b-3p target (MIR33A Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
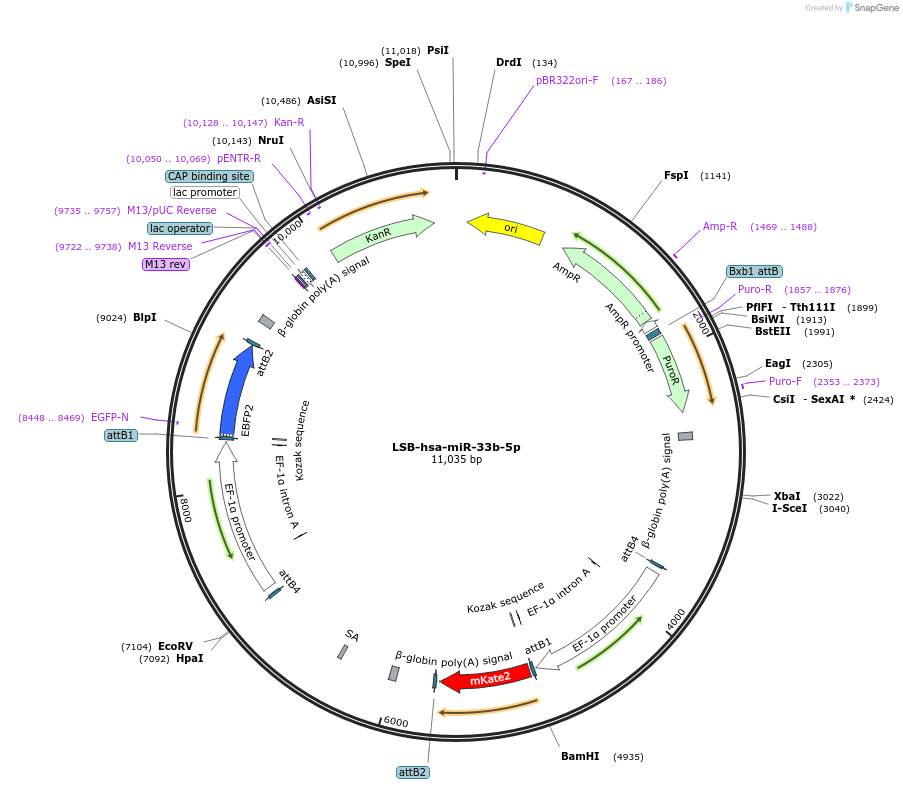
LSB-hsa-miR-33b-5p
Plasmid#103457PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-33b-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-33b-5p target (MIR33B Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
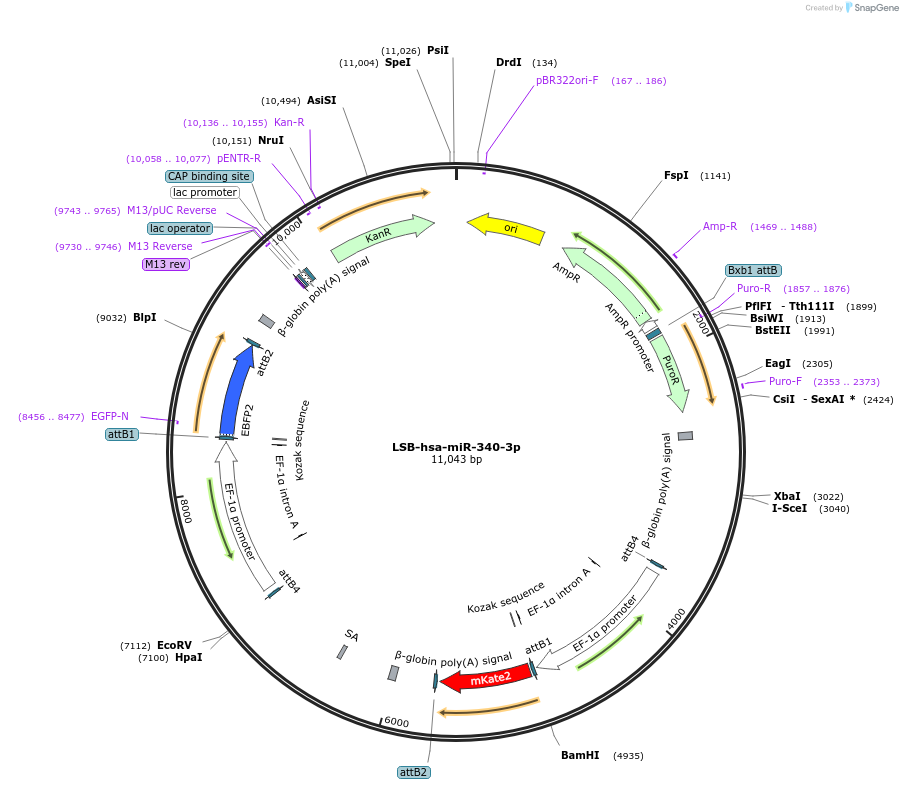
LSB-hsa-miR-340-3p
Plasmid#103458PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-340-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-340-3p target (MIR33B Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
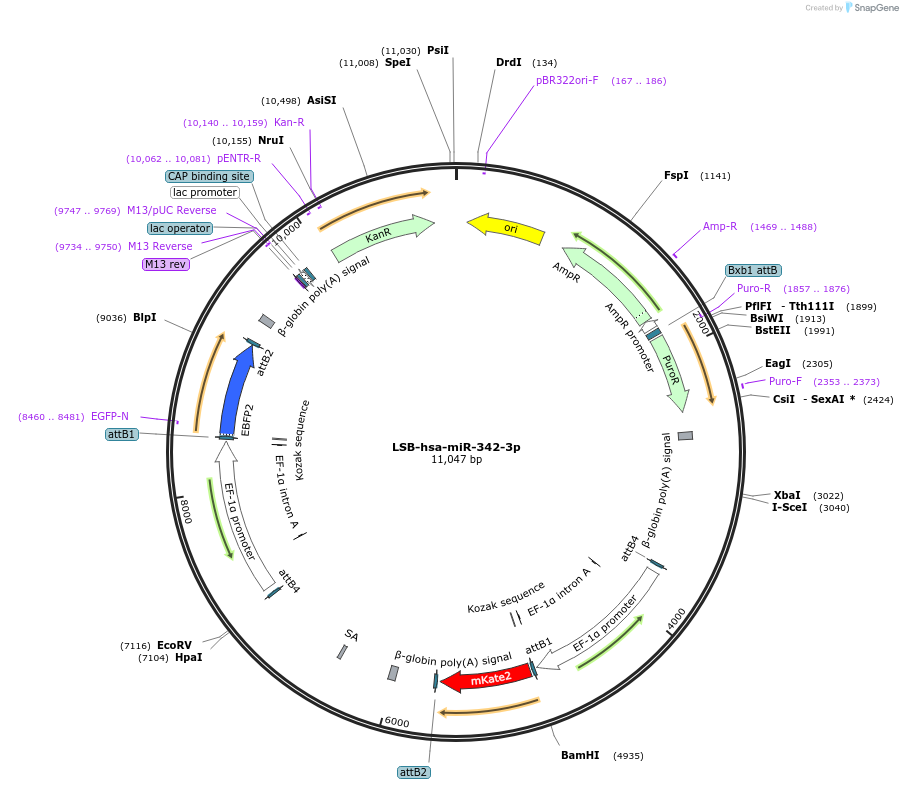
LSB-hsa-miR-342-3p
Plasmid#103460PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-342-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-342-3p target (MIR340 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
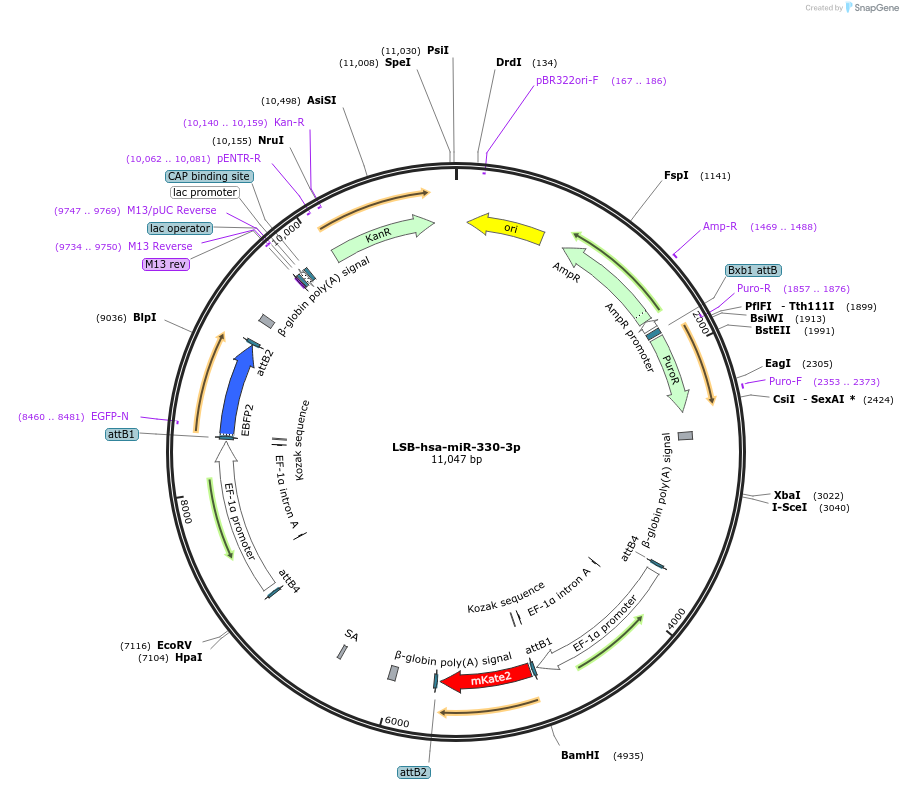
LSB-hsa-miR-330-3p
Plasmid#103442PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-330-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-330-3p target (MIR330 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
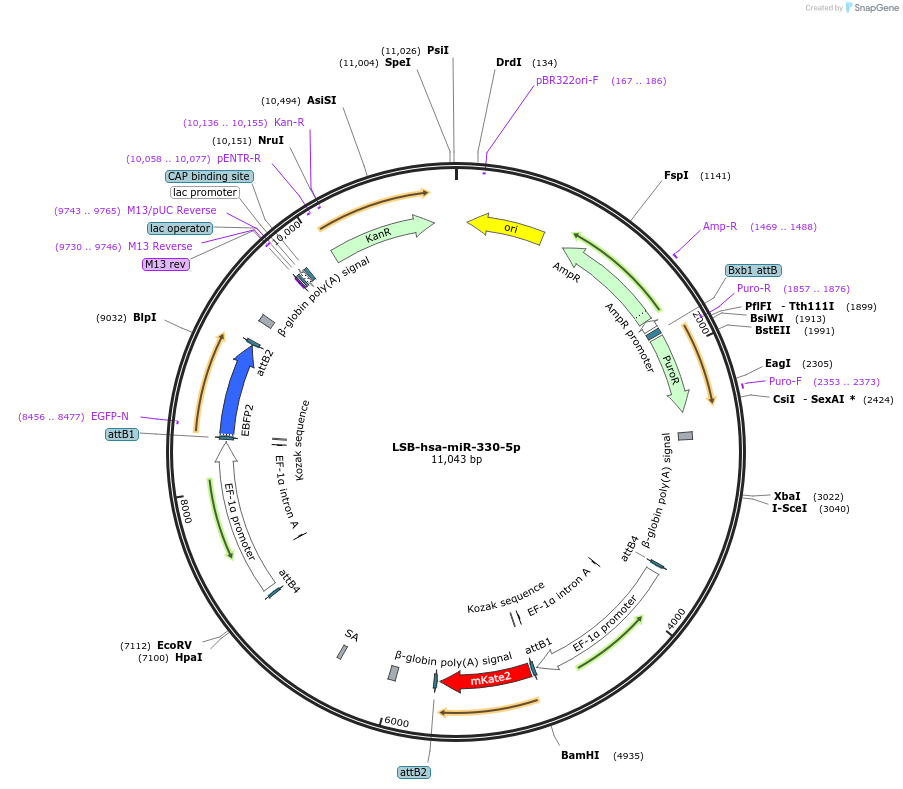
LSB-hsa-miR-330-5p
Plasmid#103443PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-330-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-330-5p target (MIR330 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
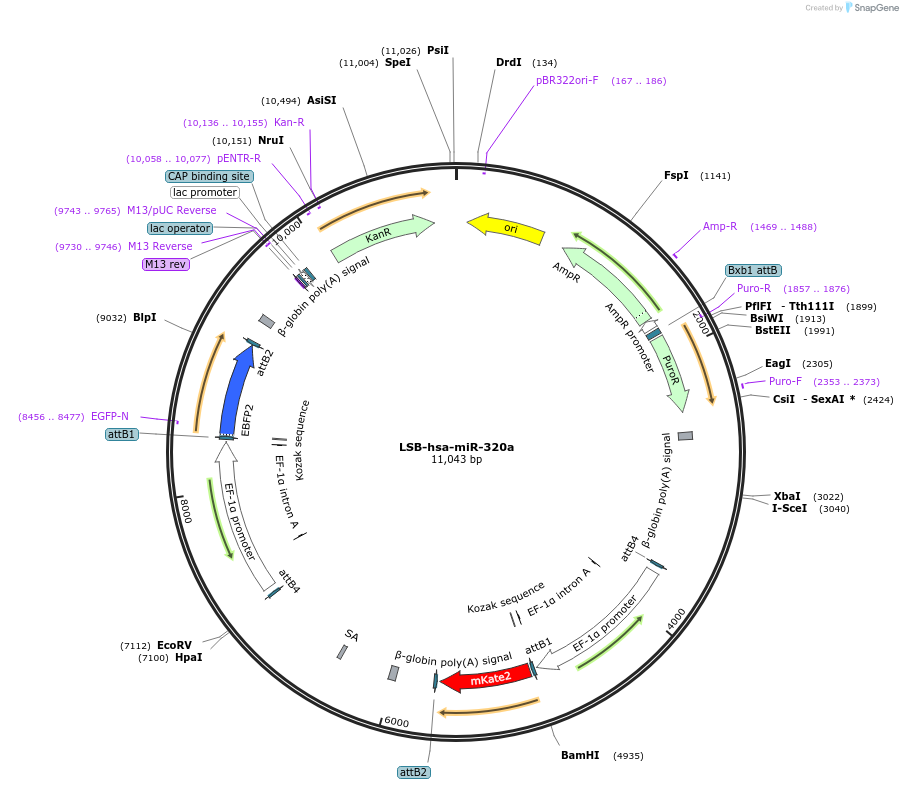
LSB-hsa-miR-320a
Plasmid#103432PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-320a target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-320a target (MIR320A Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
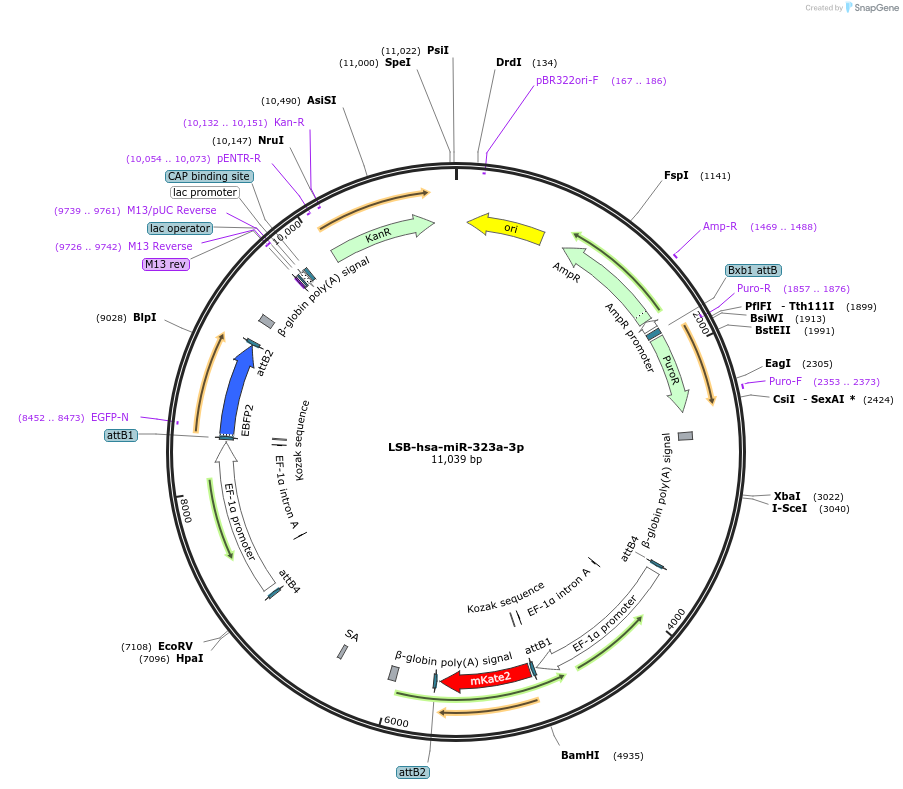
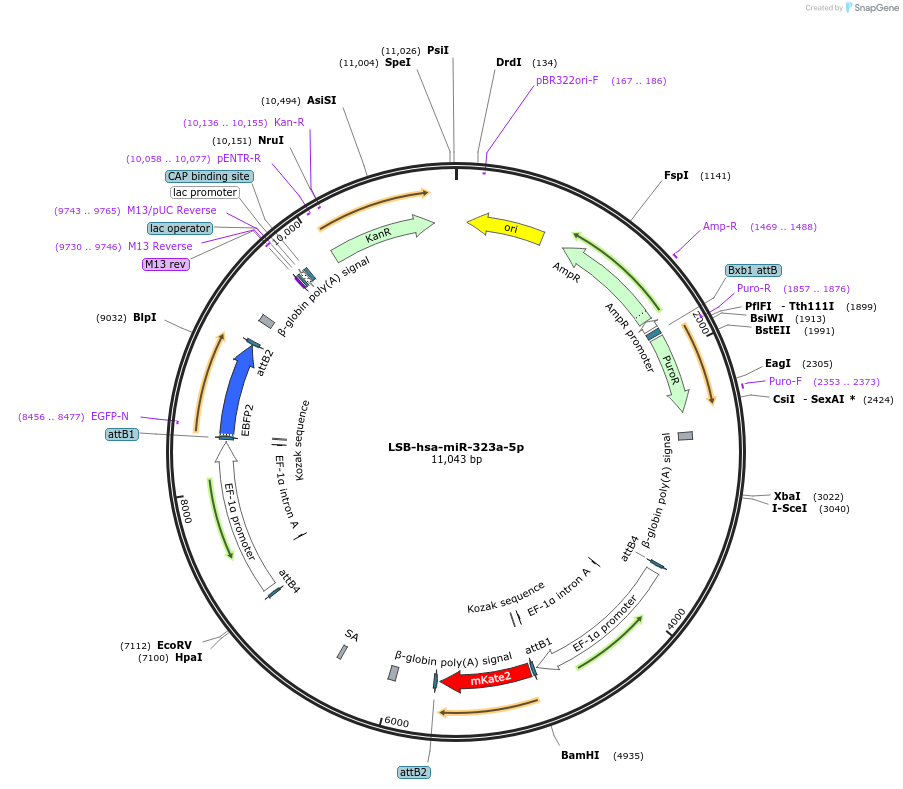
LSB-hsa-miR-323a-3p
Plasmid#103433PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-323a-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-323a-3p target (MIR323A Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
LSB-hsa-miR-323a-5p
Plasmid#103434PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-323a-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-323a-5p target (MIR323A Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
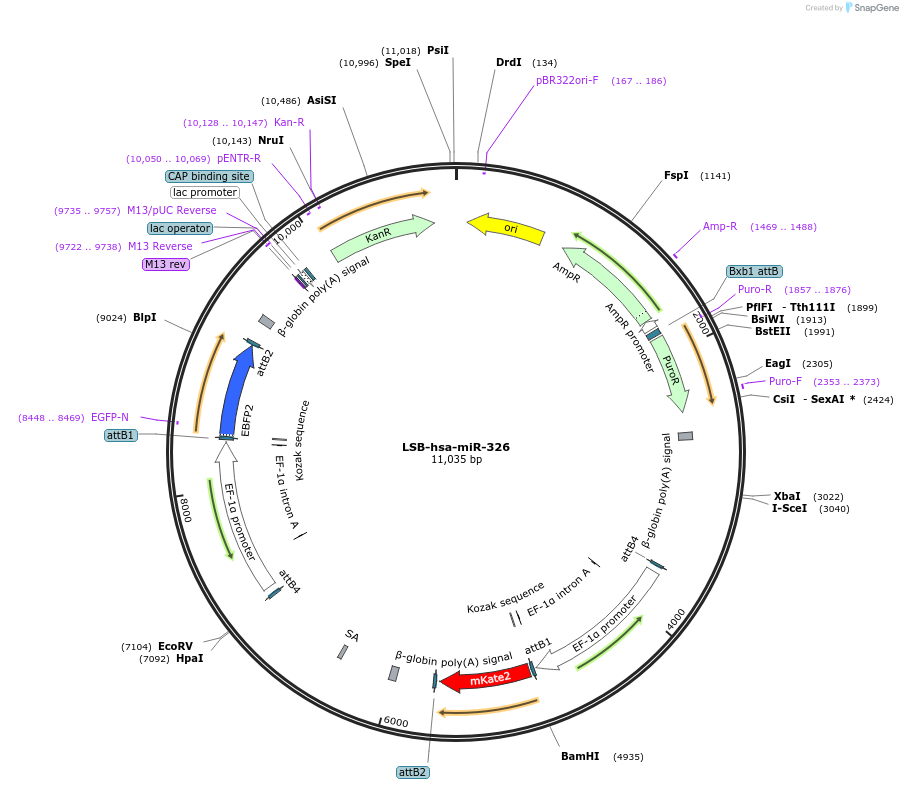
LSB-hsa-miR-326
Plasmid#103437PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-326 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
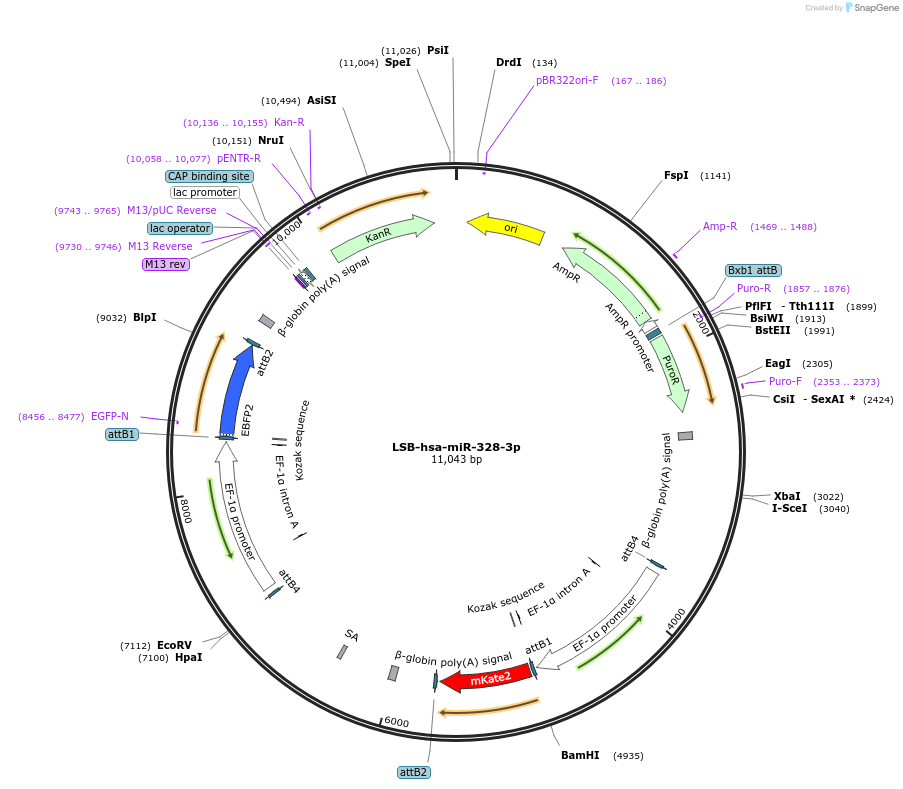
LSB-hsa-miR-328-3p
Plasmid#103438PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-328-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-328-3p target (MIR328 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
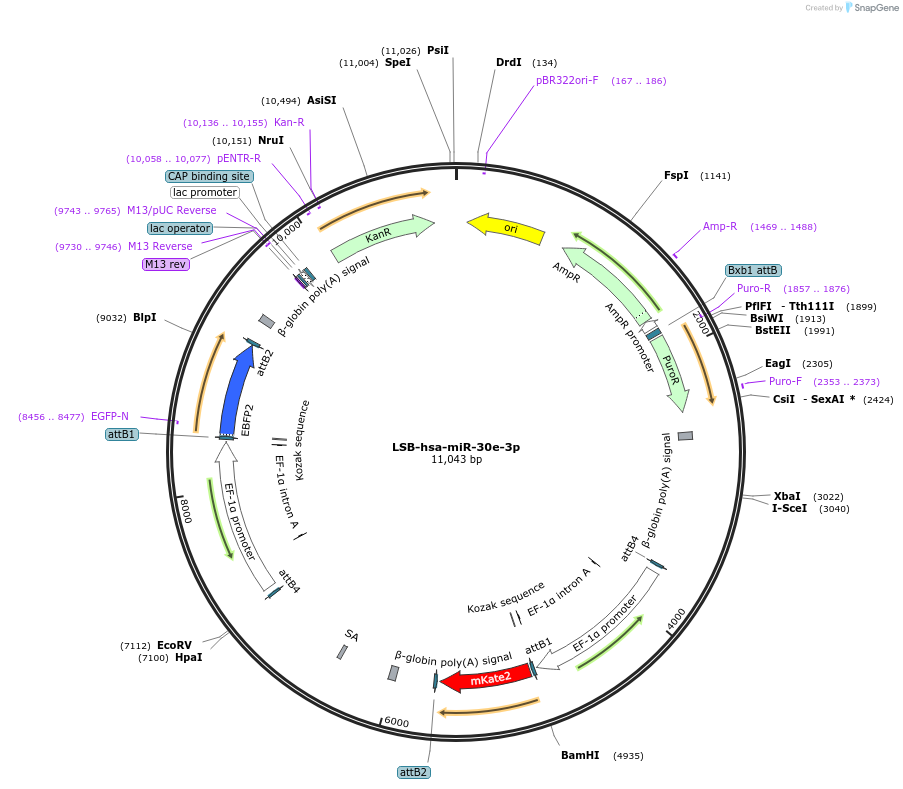
LSB-hsa-miR-30e-3p
Plasmid#103418PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-30e-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-30e-3p target (MIR30E Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
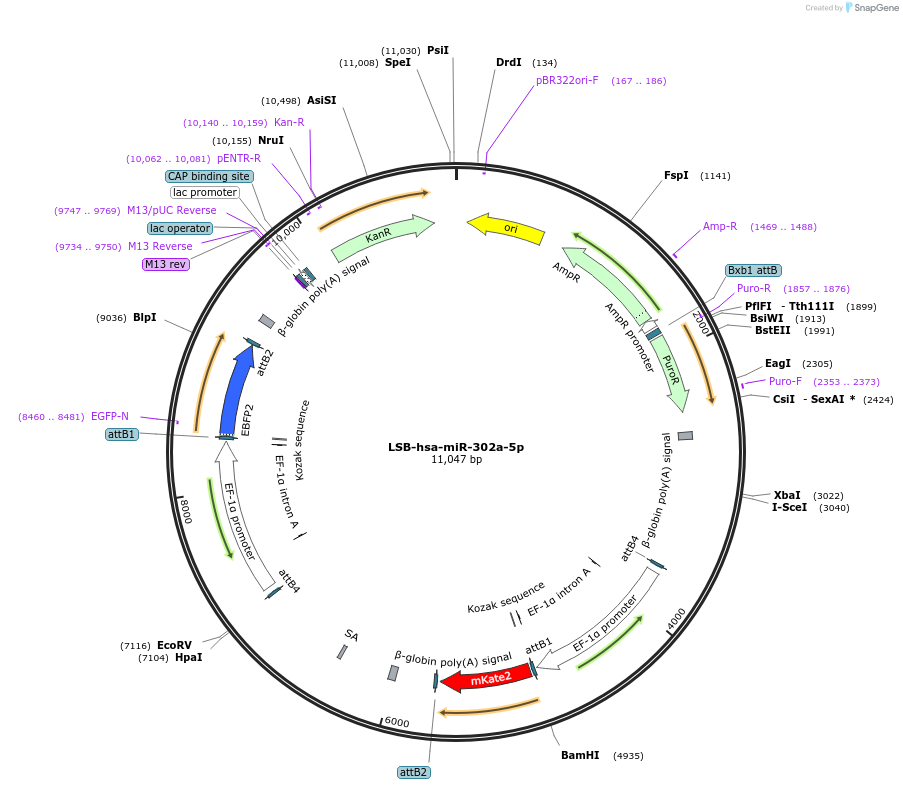
LSB-hsa-miR-302a-5p
Plasmid#103401PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-302a-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-302a-5p target (MIR302A Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
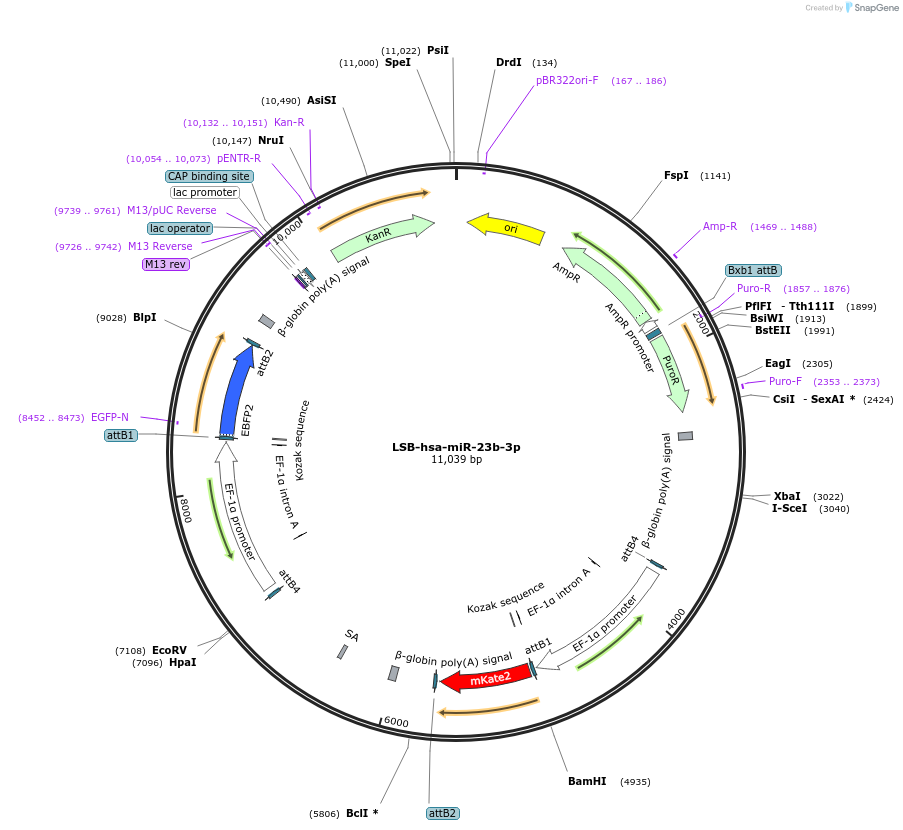
LSB-hsa-miR-23b-3p
Plasmid#103373PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-23b-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-23b-3p target (MIR23B Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
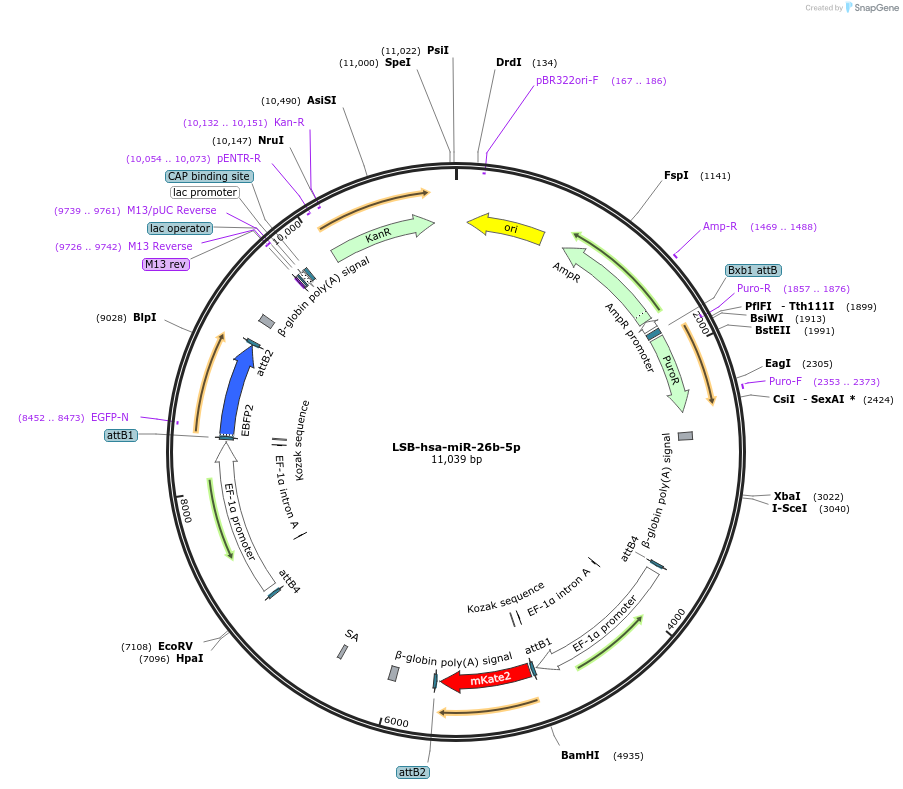
LSB-hsa-miR-26b-5p
Plasmid#103380PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-26b-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-26b-5p target (MIR26B Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
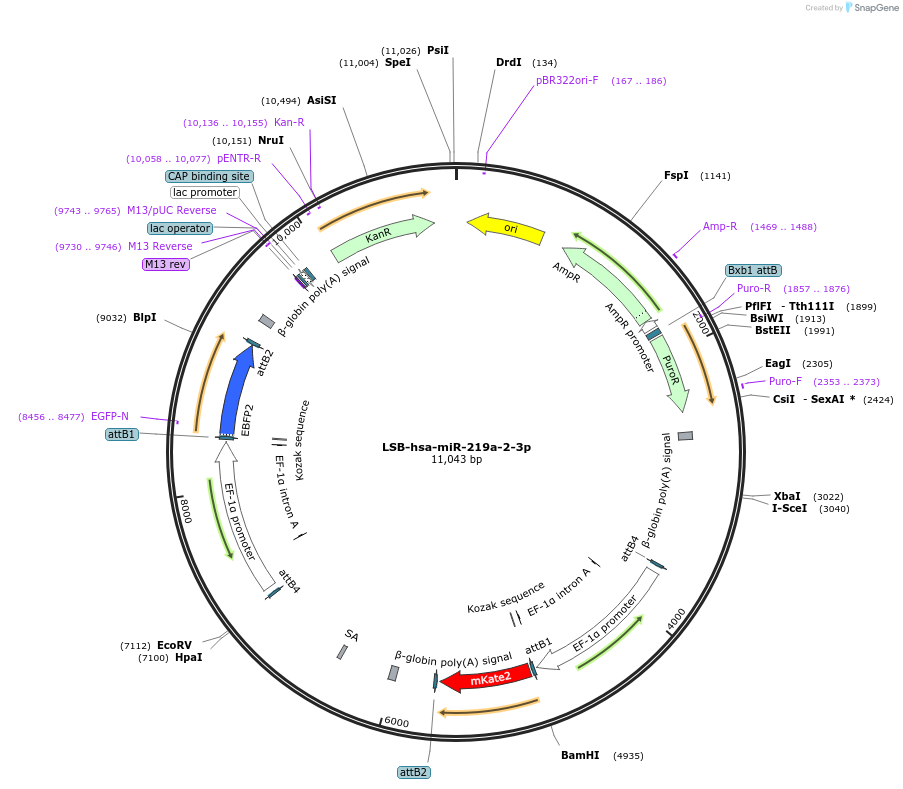
LSB-hsa-miR-219a-2-3p
Plasmid#103359PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-219a-2-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-219a-2-3p target (MIR219A2 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
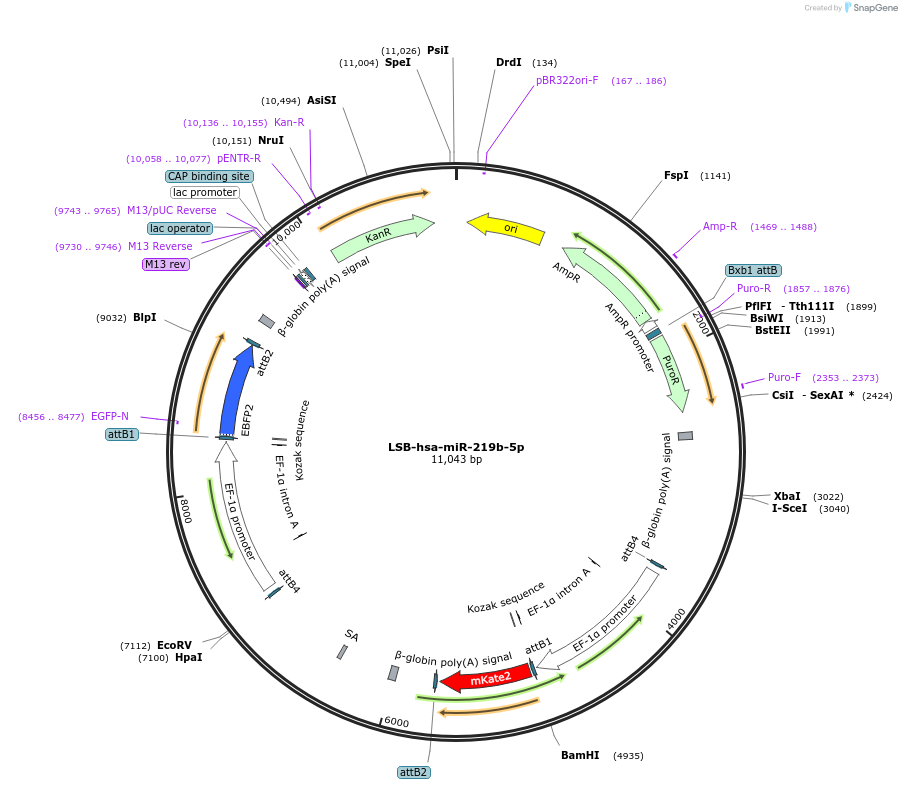
LSB-hsa-miR-219b-5p
Plasmid#103362PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-219b-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-219b-5p target (MIR219B Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only