We narrowed to 11,082 results for: CHL
-
Plasmid#84483PurposeAAV-mediated expression of ChrimsonR-tdTomato under the Syn promoter, in floxed/forward (Cre-dependent) manner. Cre turns gene off. Using bGHpA signal. tdTomato is a codon diversified version.DepositorInsertChrimsonR-tdTomato
UseAAVTagstdTomatoExpressionMammalianMutationChrimson K176R mutantPromoterhuman synapsin promoterAvailable SinceApril 26, 2017AvailabilityAcademic Institutions and Nonprofits only -
GFP-KIF1B beta-600-1400
Plasmid#79109PurposeMammalian expression of GFP-KIF1B beta-600-1400DepositorInsertGFP-KIF1B beta (600-1400 amino acids) (KIF1B Human)
TagsGFPExpressionMammalianMutationexon 25 missing (natural splice varient)PromoterCMVAvailable SinceJune 14, 2016AvailabilityAcademic Institutions and Nonprofits only -
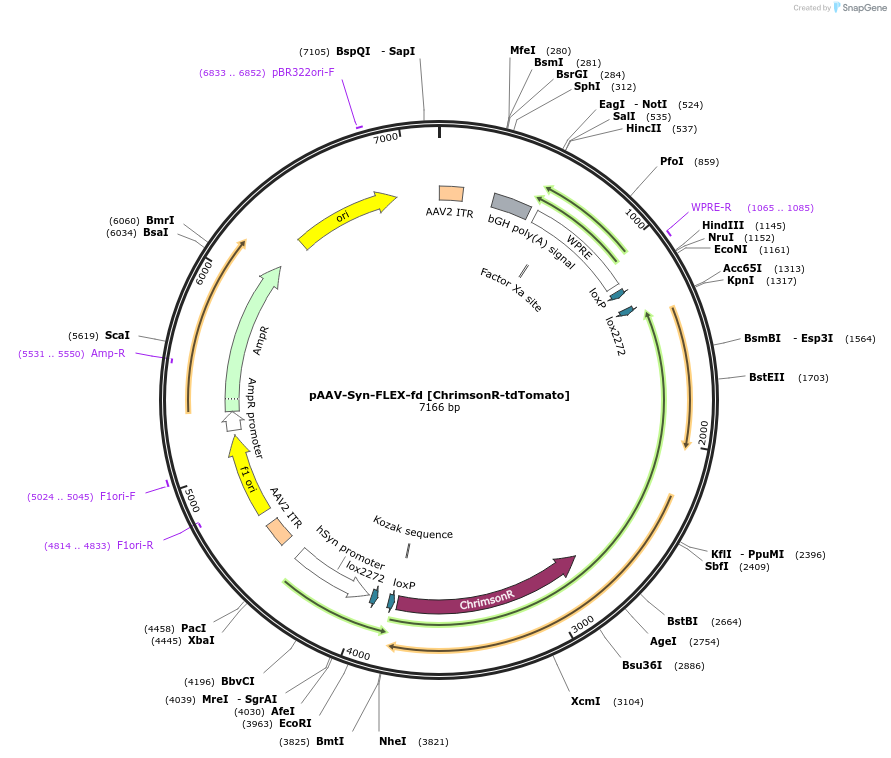
AAV-Syn-FLPX-rc [ChrimsonR-tdTomato]
Plasmid#128589PurposeAAV-mediated expression of ChrimsonR-tdTomato under the Syn promoter in frt/reversed (Flp-dependent) manner. tdTomato has codons varied to reduce recombination.DepositorHas ServiceAAV8InsertChrimsonR-tdTomato
UseAAVTagstdTomato (codon diversified version)ExpressionMammalianMutationChrimson K176R mutantPromoterSynAvailable SinceSept. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
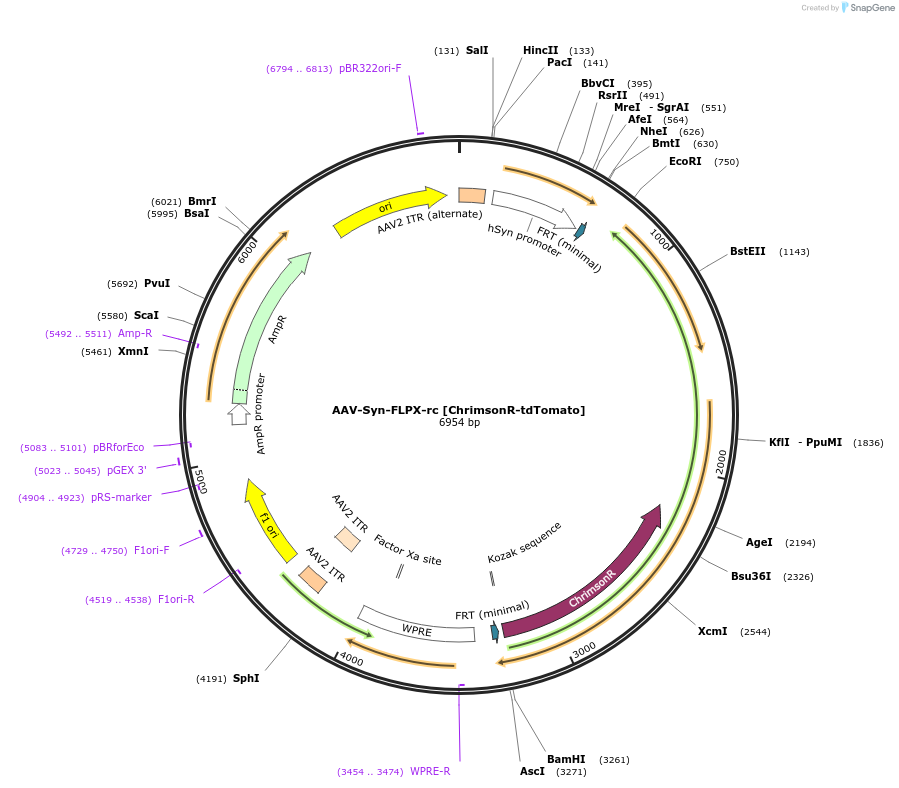
AAV-EF1α1.1-FLPX-rc [ChrimsonR-tdTomato]
Plasmid#128587PurposeAAV-mediated expression of ChrimsonR-tdTomato under the EF1α1.1 promoter in frt/reversed (Flp-dependent) manner. tdTomato has codons varied to reduce recombination.DepositorInsertChrimsonR-tdTomato
UseAAVTagstdTomato (codon diversified version)ExpressionMammalianMutationChrimson K176R mutantPromoterEF1α (1.1 kb short version)Available SinceSept. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
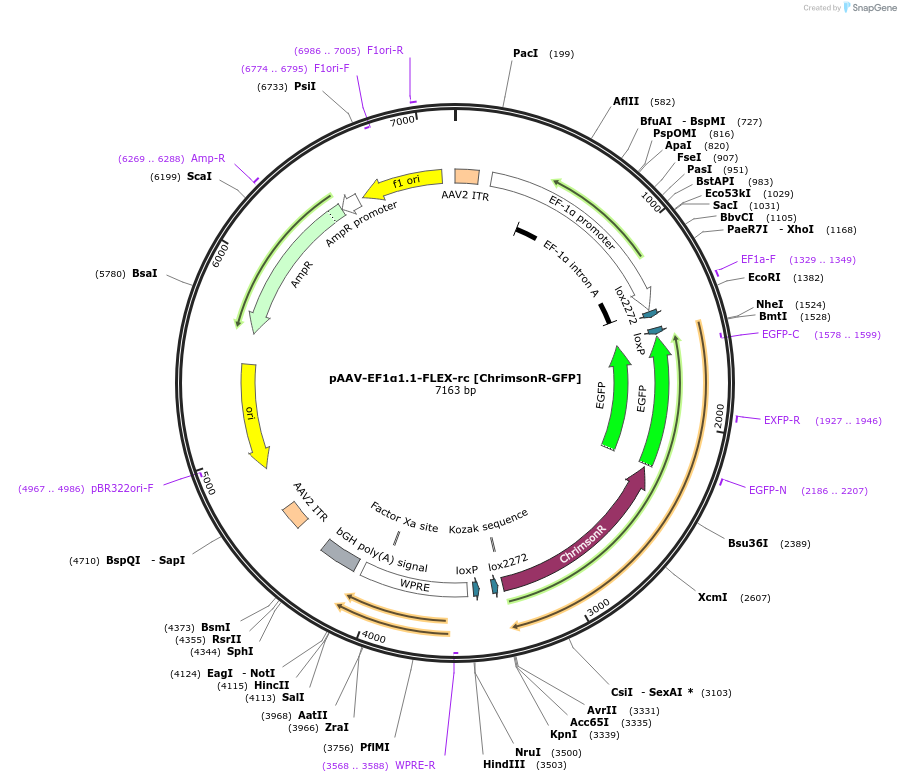
pAAV-EF1α1.1-FLEX-rc [ChrimsonR-GFP]
Plasmid#108273PurposeAAV-mediated expression of ChrimsonR-GFP under the EF1α promoter (1.1kb short version), in floxed/reversed (Cre-dependent) manner. Using bGHpA signal.DepositorInsertChrimsonR-GFP
UseAAVTagsGFPExpressionMammalianMutationChrimson K176R mutantPromoterEF1α promoter (1.1kb short version)Available SinceJune 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
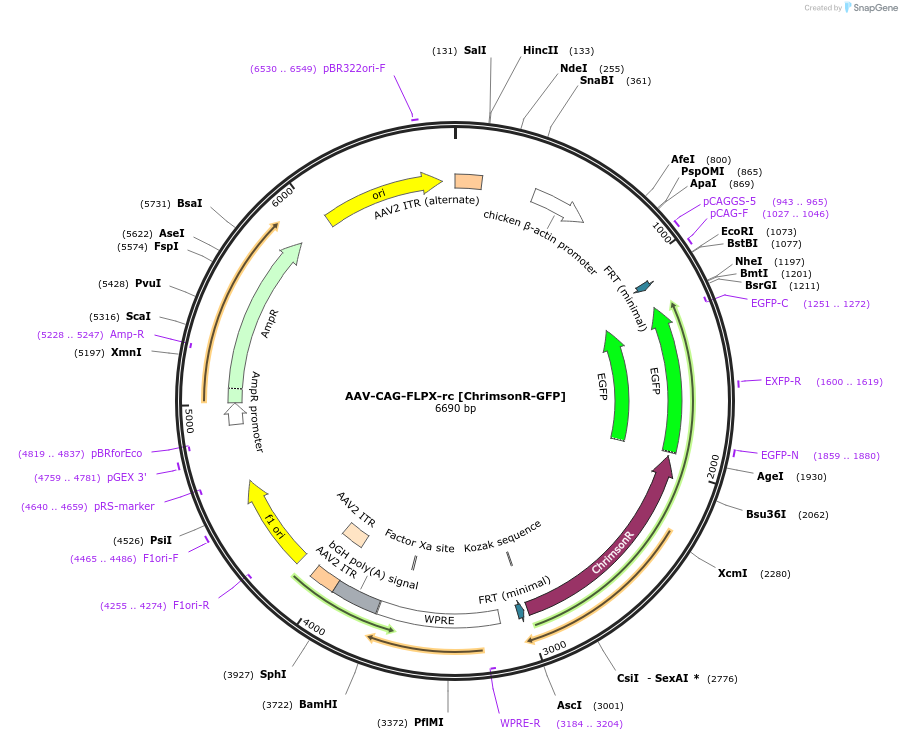
AAV-CAG-FLPX-rc [ChrimsonR-GFP]
Plasmid#118295PurposeAAV-mediated expression of ChrimsonR-GFP under the CAG promoter in frt/reversed (Flp-dependent) manner.DepositorInsertChrimsonR-GFP
UseAAVTagsGFPExpressionMammalianMutationChrimson K176R mutantPromoterCAGAvailable SinceSept. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
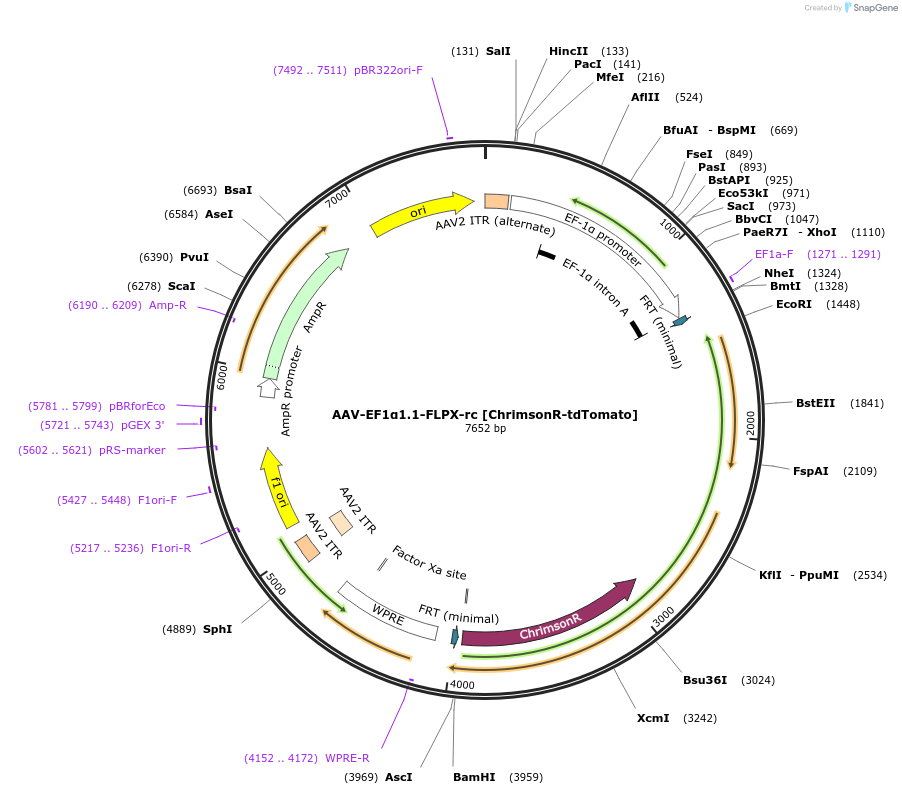
AAV-EF1α1.1-FLPX-rc [ChrimsonR-GFP]
Plasmid#128588PurposeAAV-mediated expression of ChrimsonR-GFP under the EF1α1.1 promoter in frt/reversed (Flp-dependent) manner.DepositorInsertChrimsonR-GFP
UseAAVTagsGFPExpressionMammalianMutationChrimson K176R mutantPromoterEF1α (1.1 kb short version)Available SinceSept. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
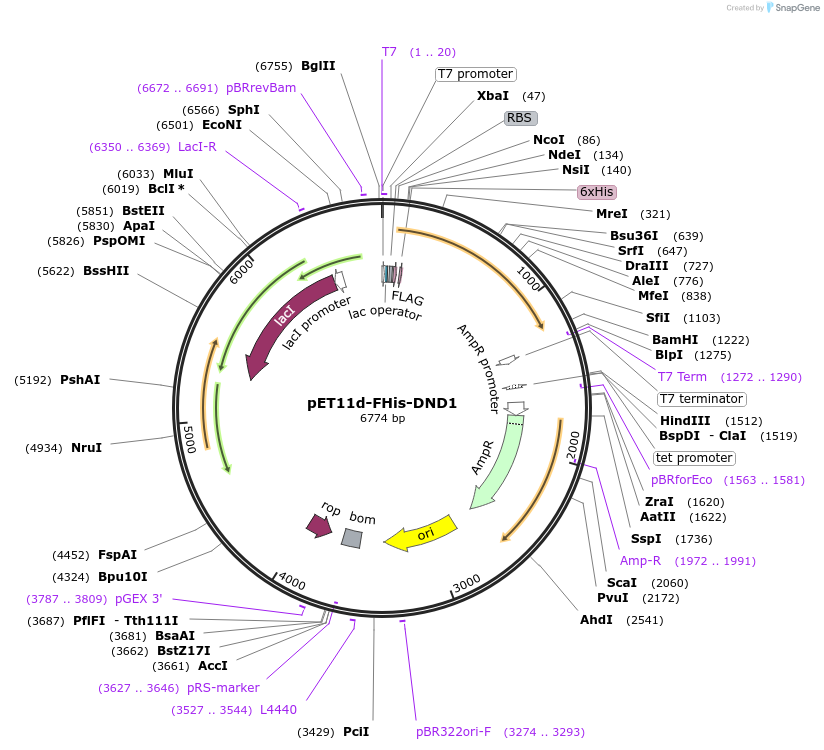
pET11d-FHis-DND1
Plasmid#70083PurposeBacterial expression of FLAG-and 6xHis-tagged human DND1 protein. Codons are optimized for bacteria expression.DepositorInsertDnd1 (DND1 Human)
Tags6xHIS and FLAGExpressionBacterialMutationCodon optimized for bacterial expressionAvailable SinceNov. 22, 2017AvailabilityAcademic Institutions and Nonprofits only -
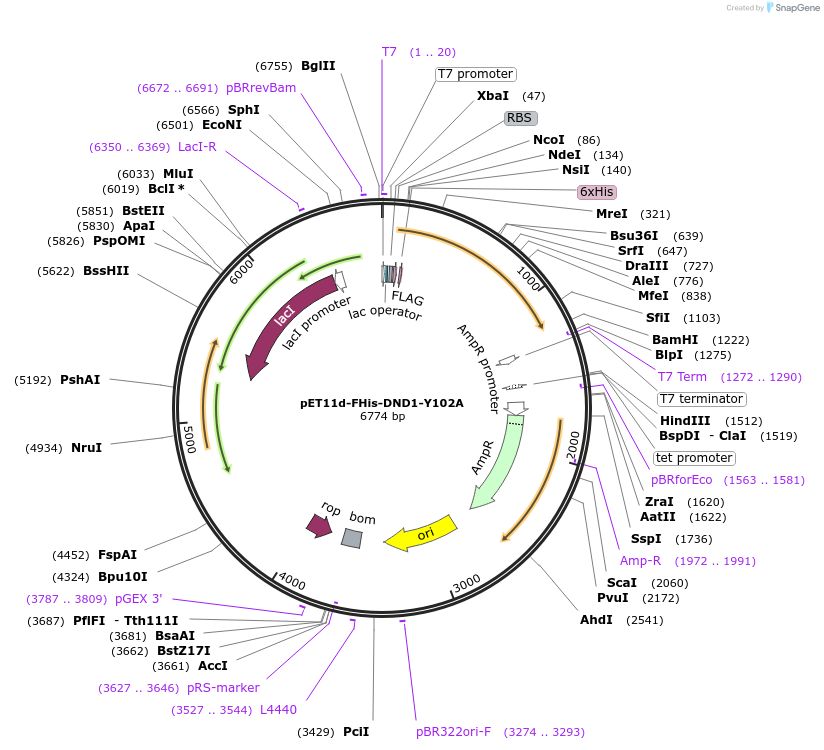
pET11d-FHis-DND1-Y102A
Plasmid#70082PurposeBacterial expression of FLAG-and 6xHis-tagged human DND1 protein with defective RRM (Y102A). Codons are optimized for bacteria expression.DepositorInsertDnd1 (DND1 Human)
Tags6xHIS and FLAGExpressionBacterialMutationY102A. Codon optimized for bacterial expressionAvailable SinceNov. 22, 2017AvailabilityAcademic Institutions and Nonprofits only -
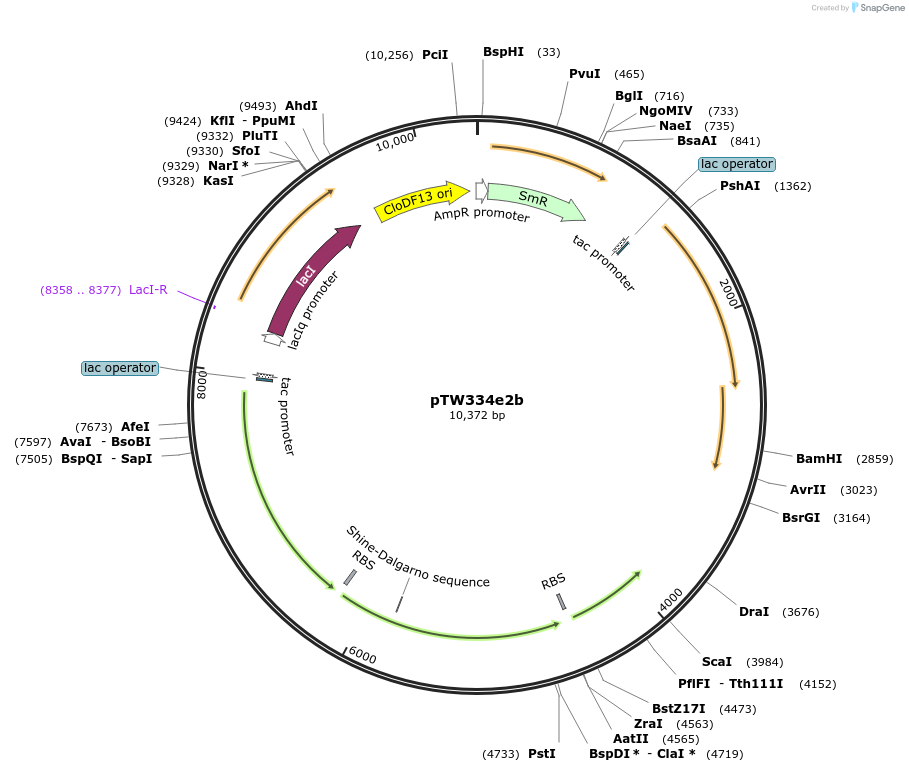
pTW334e2b
Plasmid#246587Purposesequential knock out of A. thaliana Rubisco assembly factorsDepositorInsertExpressionBacterialMutationAtBSD2W108A/L109EAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
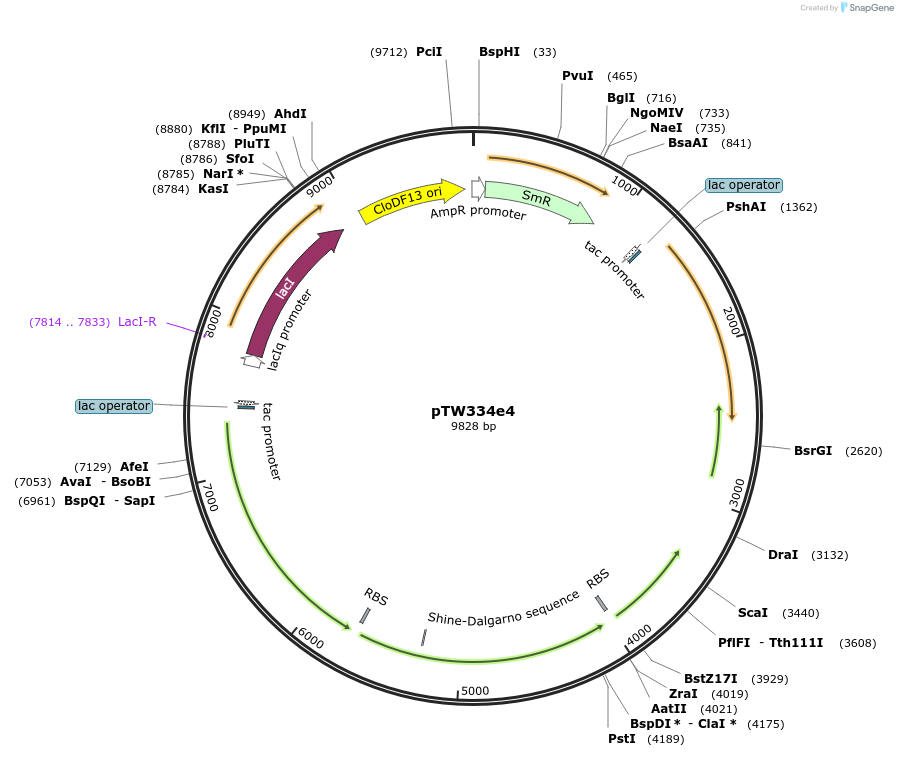
pTW334e4
Plasmid#246589Purposesequential knock out of A. thaliana Rubisco assembly factorsDepositorInsertExpressionBacterialAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
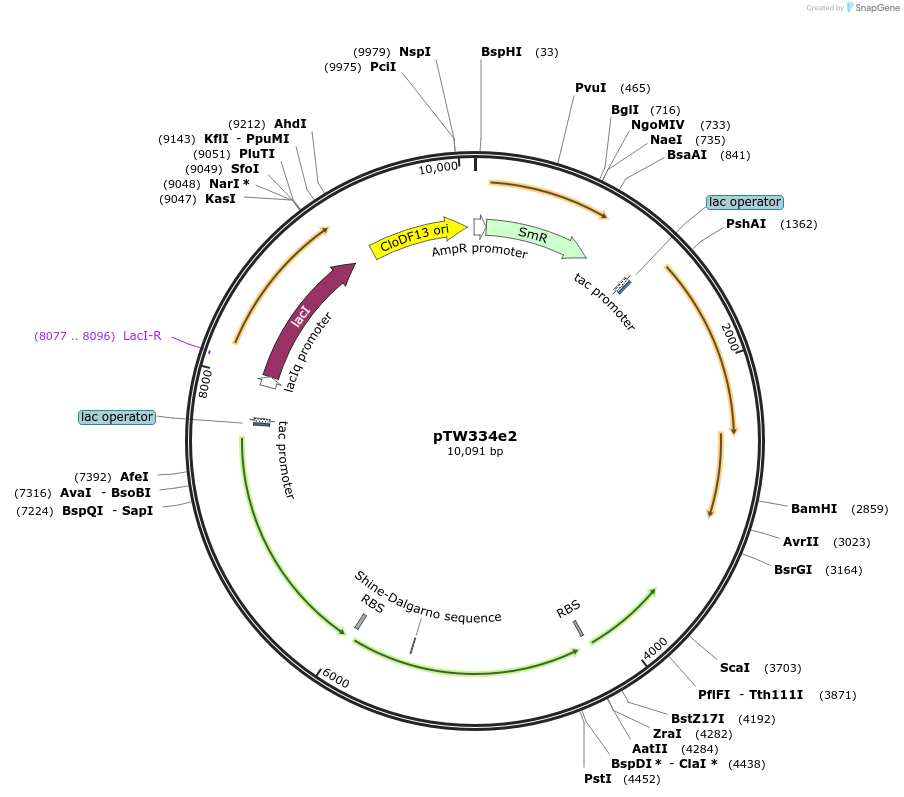
pTW334e2
Plasmid#246586Purposesequential knock out of A. thaliana Rubisco assembly factorsDepositorInsertExpressionBacterialAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
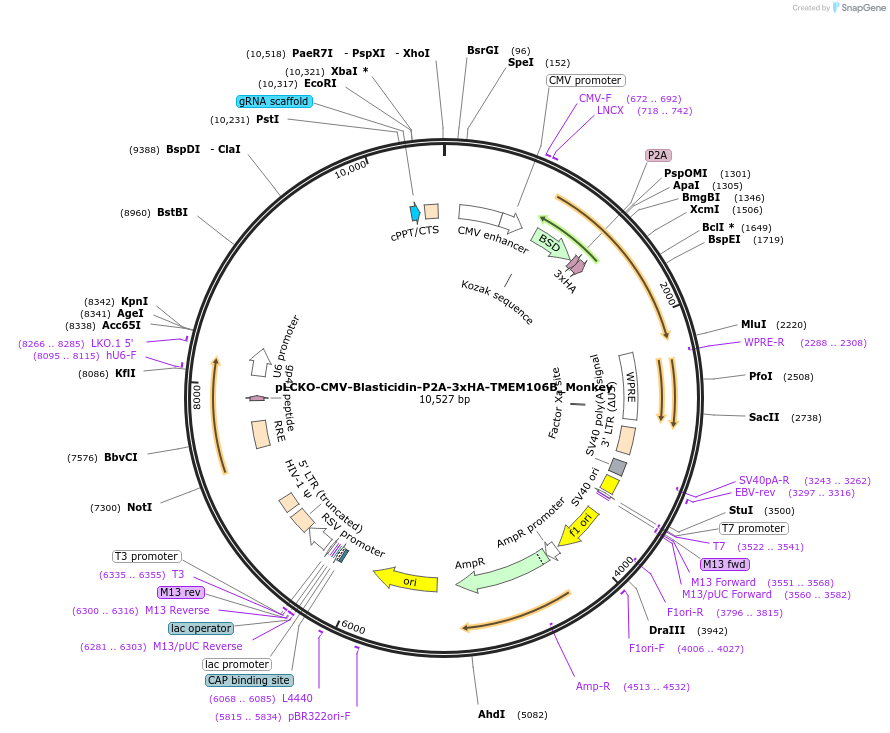
pLCKO-CMV-Blasticidin-P2A-3xHA-TMEM106B_Monkey
Plasmid#238132PurposeThis plasmid encodes the full-length African Green Monkey (Chlorocebus sabaeus) TMEM106B protein fused with a 3xHA tag, along with an N-terminal blasticidin selection marker.DepositorInsertTMEM106B
UseLentiviralTagsBlasticidin-P2A-3xHAExpressionMammalianPromoterCMVAvailable SinceJune 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
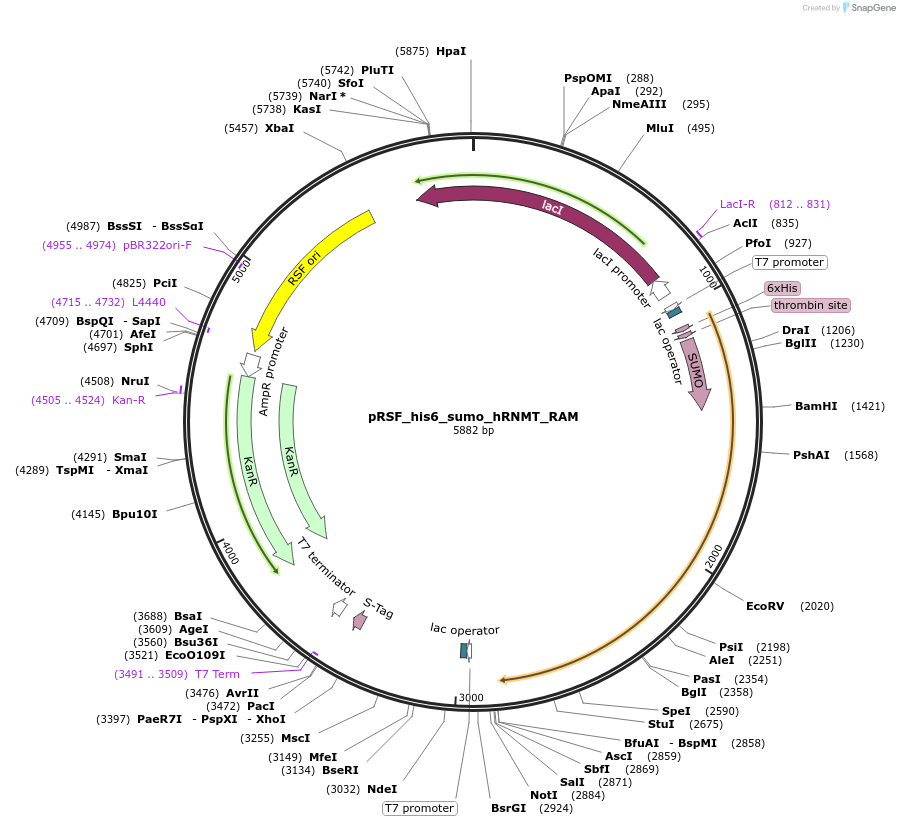
pRSF_his6_sumo_hRNMT_RAM
Plasmid#213528PurposeExpresses the human RNMT-RAM complexDepositorAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
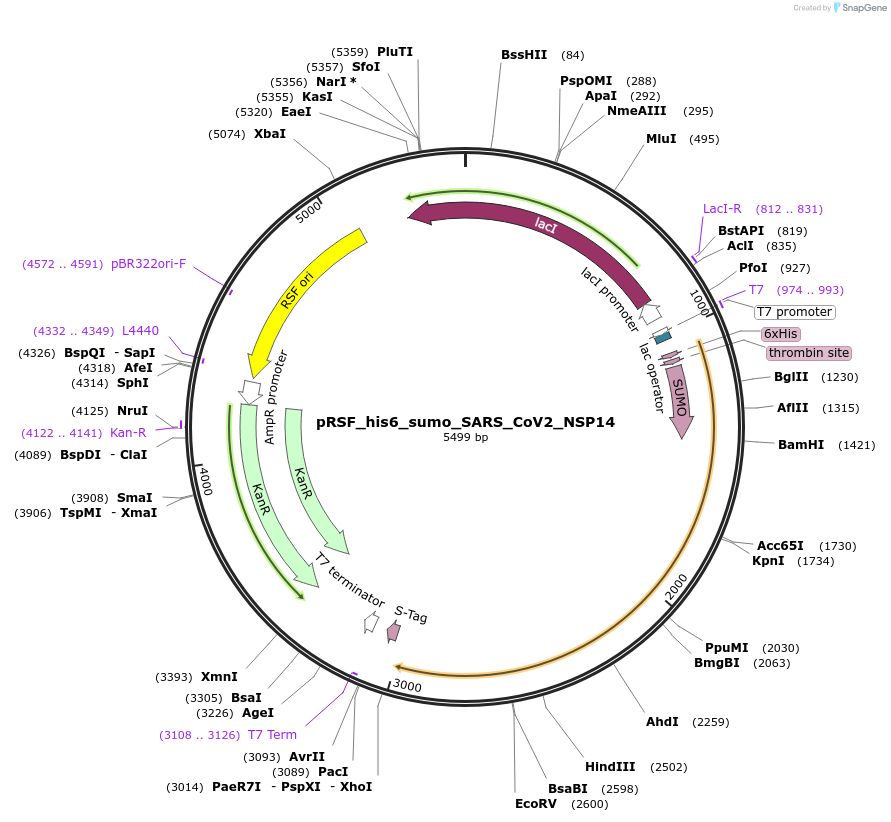
pRSF_his6_sumo_SARS_CoV2_NSP14
Plasmid#213518PurposeExpresses NSP14 of SARS-CoV-2DepositorAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
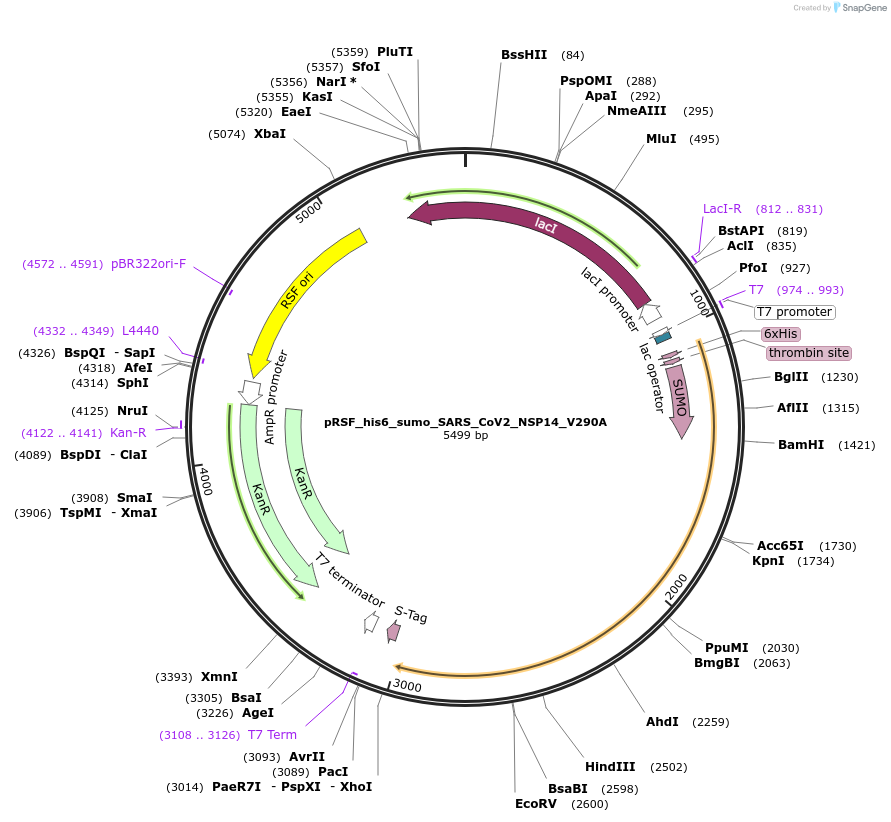
pRSF_his6_sumo_SARS_CoV2_NSP14_V290A
Plasmid#213522PurposeExpresses NSP14 of SARS-CoV-2 with a single point mutation V290ADepositorInsertSARS-CoV-2 NSP14 (ORF1ab SARS-CoV-2)
TagsHis6-sumoExpressionBacterialMutationchanged V290 to AAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
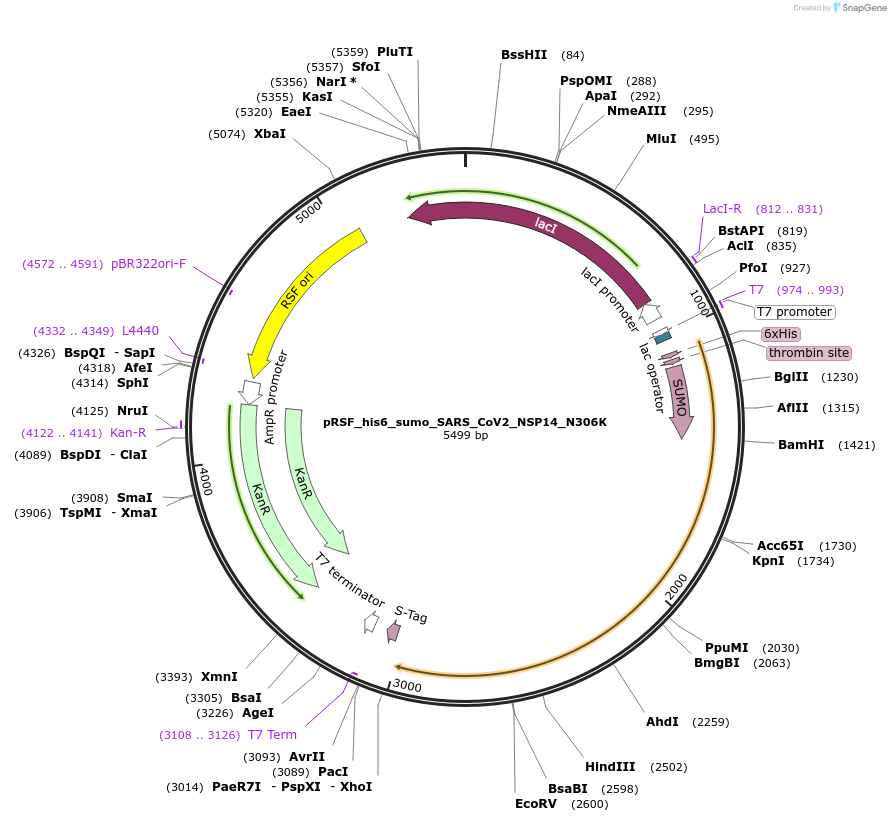
pRSF_his6_sumo_SARS_CoV2_NSP14_N306K
Plasmid#213523PurposeExpresses NSP14 of SARS-CoV-2 with a single point mutation N306KDepositorInsertSARS-CoV2-NSP14 (ORF1ab SARS-CoV-2)
TagsHis6-sumoExpressionBacterialMutationchanged N306 to KAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
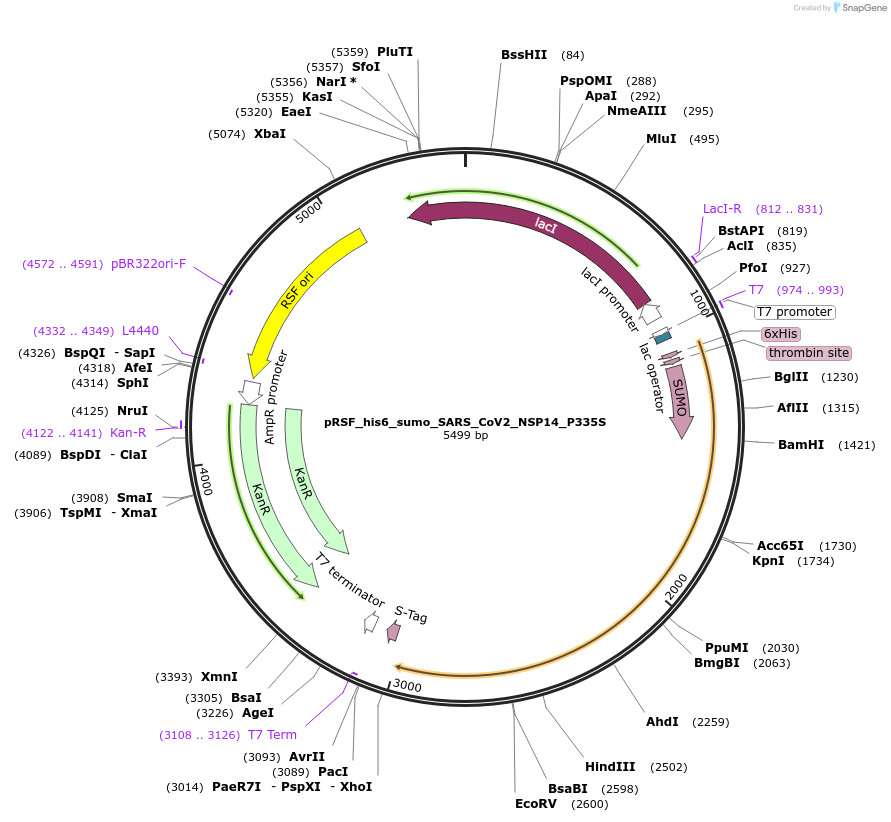
pRSF_his6_sumo_SARS_CoV2_NSP14_P335S
Plasmid#213524PurposeExpresses NSP14 of SARS-CoV-2 with a single point mutation P335SDepositorInsertSARS-CoV-2 NSP14 (ORF1ab SARS-CoV-2)
TagsHis6-sumoExpressionBacterialMutationchanged P335 to SAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
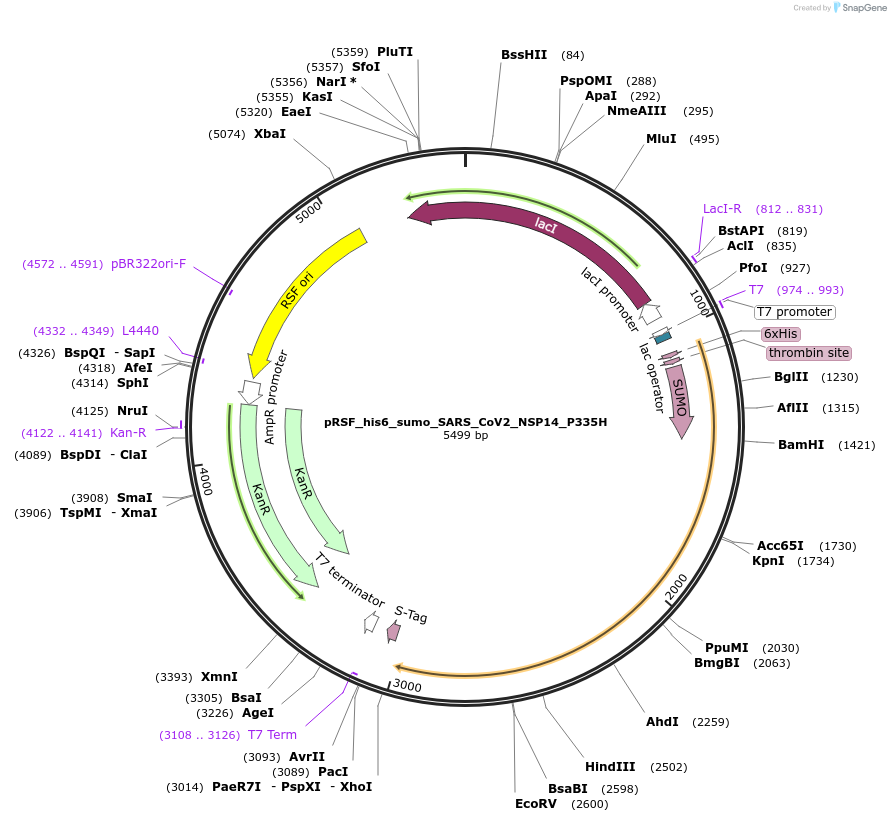
pRSF_his6_sumo_SARS_CoV2_NSP14_P335H
Plasmid#213525PurposeExpresses NSP14 of SARS-CoV-2 with a single point mutation P335HDepositorInsertSARS-CoV-2 NSP14 (ORF1ab SARS-CoV-2)
TagsHis6-sumoExpressionBacterialMutationchanged P335 to HAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
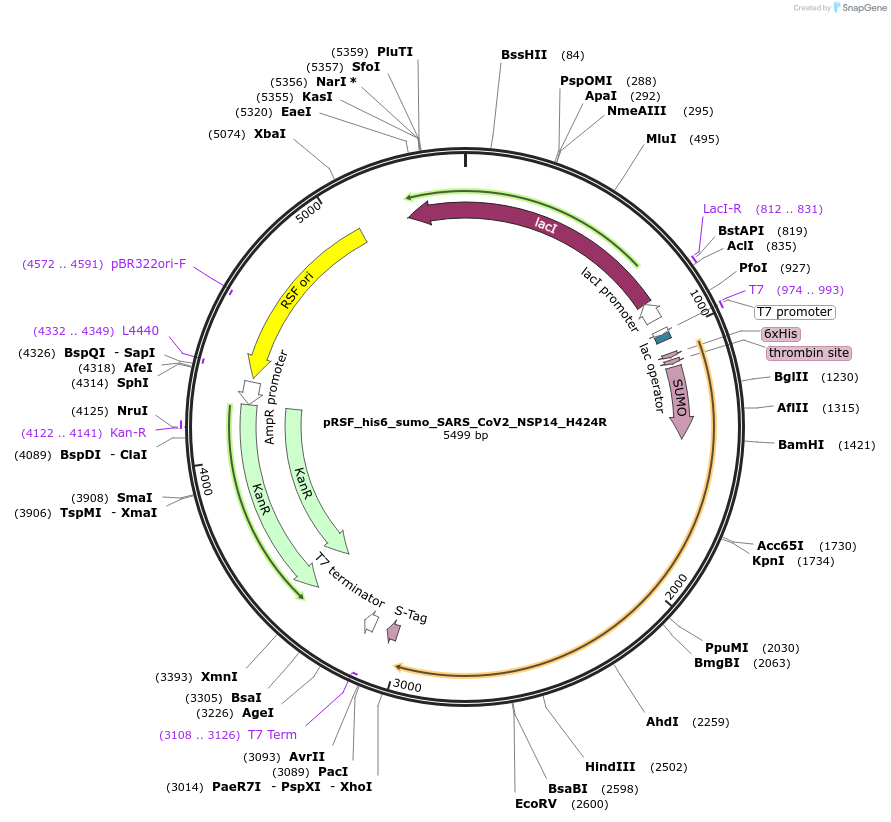
pRSF_his6_sumo_SARS_CoV2_NSP14_H424R
Plasmid#213526PurposeExpresses NSP14 of SARS-CoV-2 with a single point mutation H424RDepositorInsertSARS-CoV-2 NSP14 (ORF1ab SARS-CoV-2)
TagsHis6-sumoExpressionBacterialMutationchanged H424 to RAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only