-
PurposeSingle-wavelength extracellular glutamate sensor constructed from E. coli Gltl and cpGFP. Membrane-displayed. ** A newer version of this sensor is available. Please see iGluSnFR3 plasmids at https://www.addgene.org/browse/article/28220233/ **
-
Depositing Lab
-
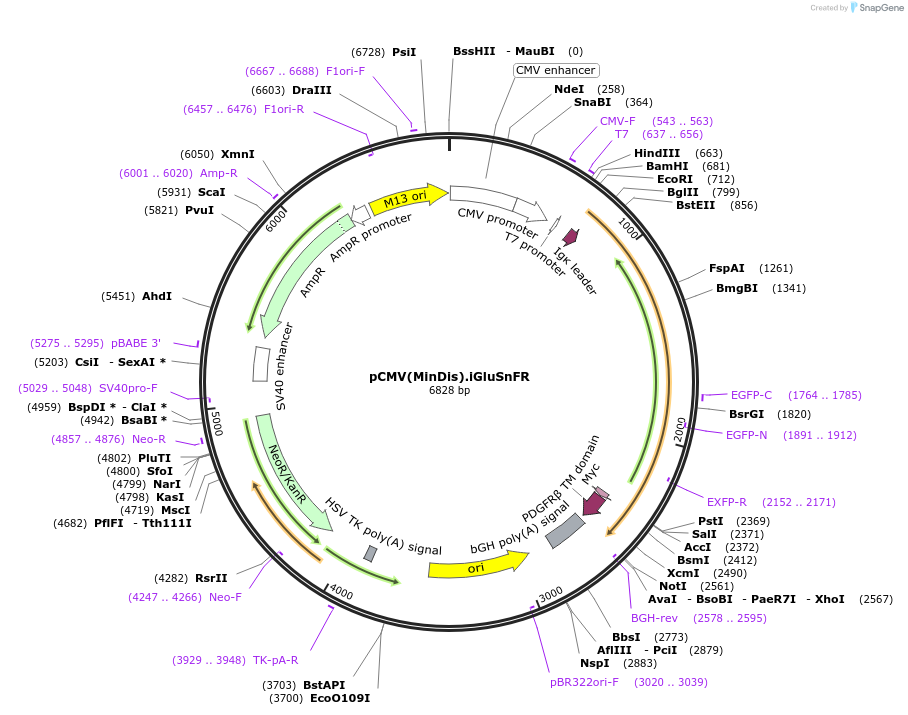
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 41732 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepCMV(MinDis) [variant of pDisplay]
-
Backbone manufacturermodified from Invitrogen
- Backbone size w/o insert (bp) 5300
- Total vector size (bp) 6829
-
Modifications to backboneFrom pDisplay: Hemagglutinin (HA) tag removed
-
Vector typeMammalian Expression ; Glutamate Biosensor
-
Selectable markersNeomycin (select with G418)
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameGltI based Glutamate Biosensor
-
Alt nameiGluSnFR
-
Alt namegltI/ybeJ
-
Alt namecpGFP146
-
SpeciesSynthetic; E. coli
-
Insert Size (bp)1570
-
MutationcpGFP146 variant from EcMBP165-cpGFP.PPYF.pRSET inserted between AA253-254 of E. coli gltI/ybeJ
- Promoter CMV
-
Tags
/ Fusion Proteins
- mouse Ig κ-chain (N terminal on backbone)
- Myc (C terminal on backbone)
- PDGFR transmembrane helix (aa 513-561) (C terminal on backbone)
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site BglII (not destroyed)
- 3′ cloning site PstI (not destroyed)
- 5′ sequencing primer CMV-F
- 3′ sequencing primer BGH-rev
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byE. coli glutamate-binding protein (gltI) from FLIP-E (Okumoto, et al. 2005, PNAS, PMID: 15939876) and cpGFP146 variant from EcMBP165-cpGFP.PPYF.pRSET (Addgene plasmid #33372)
-
Articles Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
** A newer version of this sensor is available. Please see iGluSnFR3 plasmids at https://www.addgene.org/browse/article/28220233/ **
Biosensor iGluSnFRa is a single-wavelength glutamate sensor constructed from E. coli GltI and cpGFP. iGluSnFR is bright and photostable, with 4.5 (ΔF/F)max in vitro, under both 1- and 2-photon illumination.
pCMV(MinDis).iGluSnFR was constructed by subcloning the GltI253-cpGFP.L1LV/L2NP gene from pRSET.GltI253-cpGFP.L1LV/L2NP (Addgene plasmid #41733) into the pDisplay vector (Invitrogen) between the BglII and PstI sites. The hemagglutinin (HA) tag was removed by PCR, generating the backbone variant dubbed pCMV(MinDis).
This construct encodes an N-terminal mouse immunoglobin κ-chain leader sequence, which directs the protein to the secretory pathway; the GltI253.L1LV/L2NP protein; a myc epitope tag; and at the C-terminus, a platelet derived growth factor receptor transmembrane helix, which anchors the protein to the plasma membrane, displaying it on the extracellular side.
Detailed amino acid sequences are provided in the supplementary documents available above.
Alternate plasmid names: YbeJ253.L1LVL2NP.pMinDis; pMinDis.iGluSnFR
These iGluSnFR constructs contain a K158R (GFP numbering) mutation that is not referenced in the publication. Depositing lab has confirmed that this mutation is expected and that the plasmids function as described.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pCMV(MinDis).iGluSnFR was a gift from Loren Looger (Addgene plasmid # 41732 ; http://n2t.net/addgene:41732 ; RRID:Addgene_41732) -
For your References section:
An optimized fluorescent probe for visualizing glutamate neurotransmission. Marvin JS, Borghuis BG, Tian L, Cichon J, Harnett MT, Akerboom J, Gordus A, Renninger SL, Chen TW, Bargmann CI, Orger MB, Schreiter ER, Demb JB, Gan WB, Hires SA, Looger LL. Nat Methods. 2013 Feb;10(2):162-70. doi: 10.1038/nmeth.2333. Epub 2013 Jan 13. 10.1038/nmeth.2333 PubMed 23314171