CRISPR: Purify Genomic Loci
Identifying molecules associated with a genomic region of interest in vivo is essential to understanding locus function. Using CRISPR, researchers have expanded chromatin immunoprecipitation (ChIP) to allow purification of any genomic sequence specified by a particular gRNA.
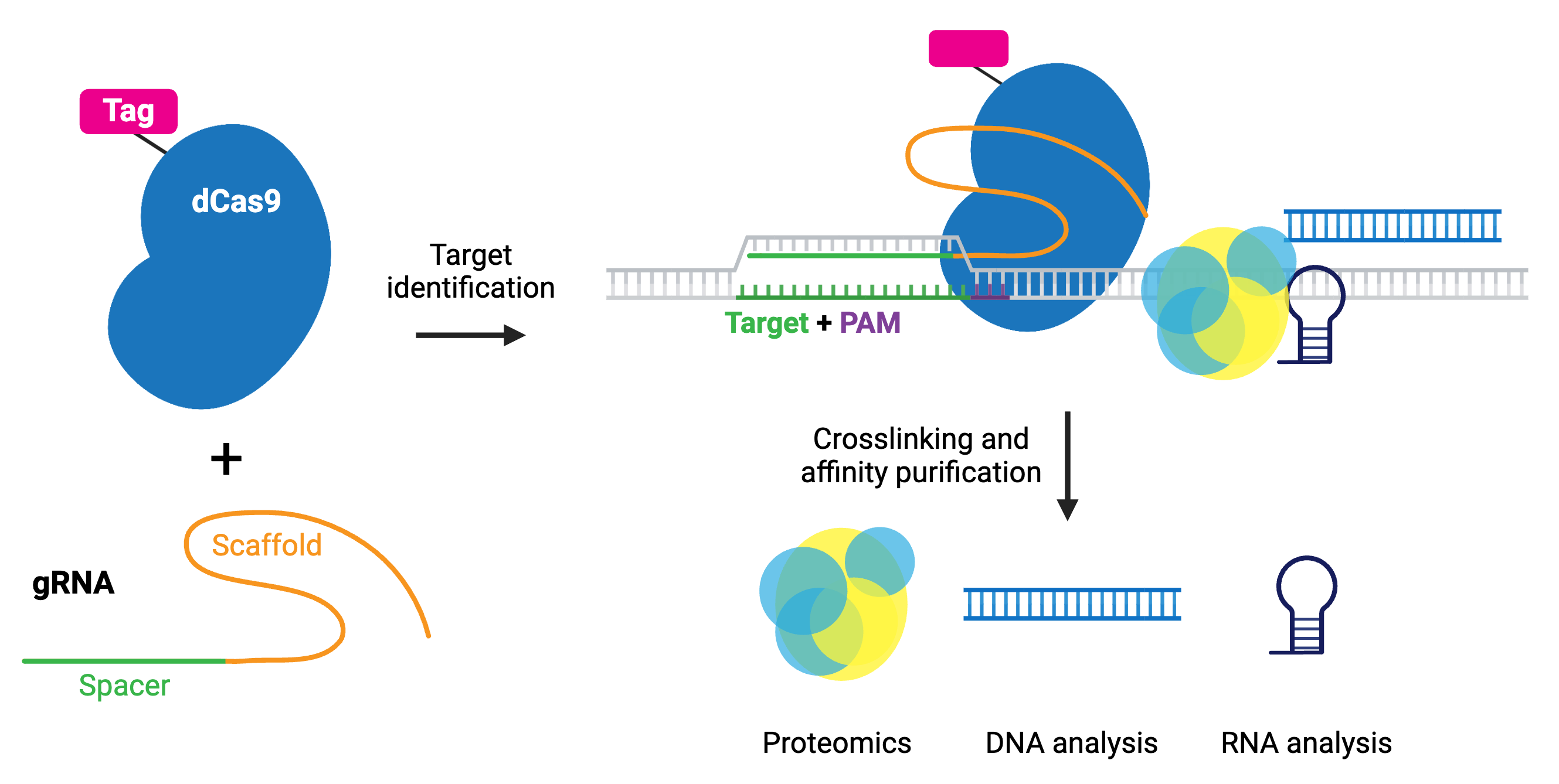
In the enChIP (engineered DNA-binding molecule-mediated ChIP) system, catalytically inactive dCas9 is used to purify genomic DNA bound by the gRNA. An epitope tag or tags can be fused to dCas9 or gRNA for efficient purification. Various epitope tags including 3xFLAG-tag, PA, and biotin tags, can be used for enChIP, as well as an anti-Cas9 antibody. Biotin tagging of dCas9 can be achieved by fusing a biotin acceptor site to dCas9 and co-expressing BirA biotin ligase, as seen in the CAPTURE system. The locus is subsequently isolated by affinity purification against the epitope tag.
Compared to conventional methods for genomic purification, CRISPR-based purification methods are more straightforward and enable direct identification of molecules associated with a genomic region of interest in vivo.
Browse, sort, or search the tables below for CRISPR plasmids designed for the purification of genomic loci. To learn more about purification using CRISPR and other CRISPR topics, read our CRISPR Guide.
Mammalian
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
Bacteria
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
Yeast
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
CRISPR Resources
Addgene has a large selection of CRISPR plasmids and resources. Find more CRISPR functions along with plasmids categorized by organism by visiting our CRISPR plasmids page. Find a comprehensive list of CRISPR resources by visiting our CRISPR reference page.
Content last reviewed: 17 October 2025
Do you have suggestions for other plasmids that should be added to this list?
Fill out our Suggest a Plasmid form or e-mail [email protected] to help us improve this resource!



