CRISPR Plasmids: Visualize Genomic Loci
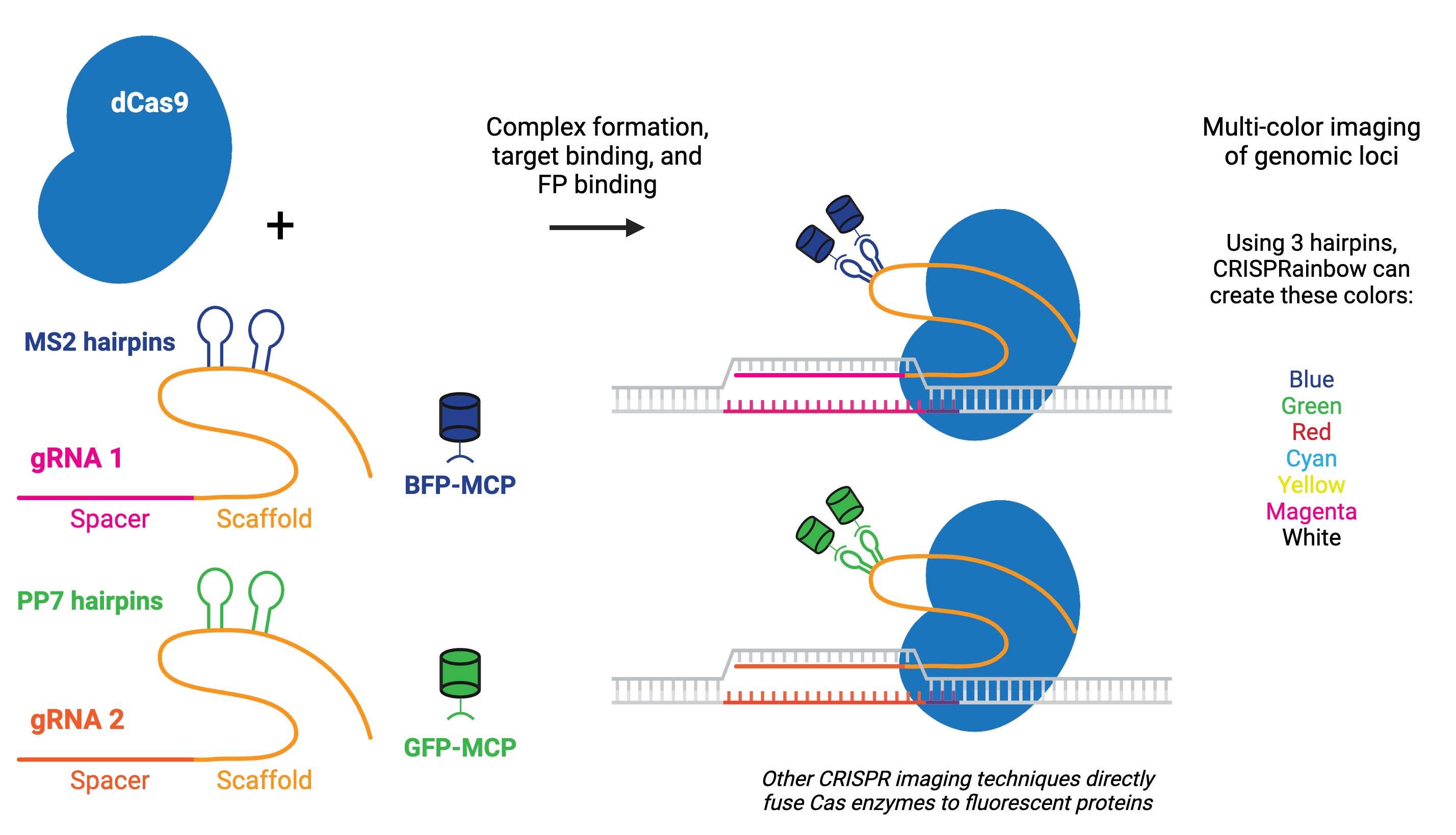
Using catalytically inactive Cas9 (dCas9) fused to a fluorescent marker like GFP, researchers have turned dCas9 into a customizable DNA labeler compatible with fluorescence microscopy in living cells. Alternatively, gRNAs can be fused to protein-interacting RNA aptamers, which recruit specific RNA-binding proteins (RBPs) tagged with fluorescent proteins to visualize targeted genomic loci.
CRISPR imaging has numerous advantages over other imaging techniques, including that it is easy to implement due to the simplicity of gRNA design, programmable for different genomic loci, capable of detecting multiple genomic loci, and compatible with live cell imaging. Compared to techniques like fluorescence in situ hybridization (FISH), CRISPR imaging offers a unique method for detecting the chromatin dynamics in living cells.
The fluorescent CRISPR system has been used for dynamic tracking of repetitive and non-repetitive genomic loci, as well as chromosome painting in living cells. Visualizing a specific genomic locus requires recruitment of many copies of labeled proteins to the given region. For example, chromosome-specific repetitive loci can be efficiently visualized in living cells using a single gRNA that has multiple targeted sequences in close proximity. A non-repetitive genomic locus can also be labeled by co-delivering multiple gRNAs that tile the locus. Chromosome painting requires delivery of hundreds of gRNAs with target sites throughout the chromosome.
Browse, sort, or search the tables below for CRISPR plasmids designed for the visualization of genomic loci. To learn more about visualization using CRISPR and other CRISPR topics, read our CRISPR Guide.
Mammalian
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
CRISPR Resources
Addgene has a large selection of CRISPR plasmids and resources. Find more CRISPR functions along with plasmids categorized by organism by visiting our CRISPR plasmids page. Find a comprehensive list of CRISPR resources by visiting our CRISPR reference page.
Content last reviewed: 17 October 2025
Do you have suggestions for other plasmids that should be added to this list?
Fill out our Suggest a Plasmid form or e-mail [email protected] to help us improve this resource!



