E11 Bio PRISM Collection
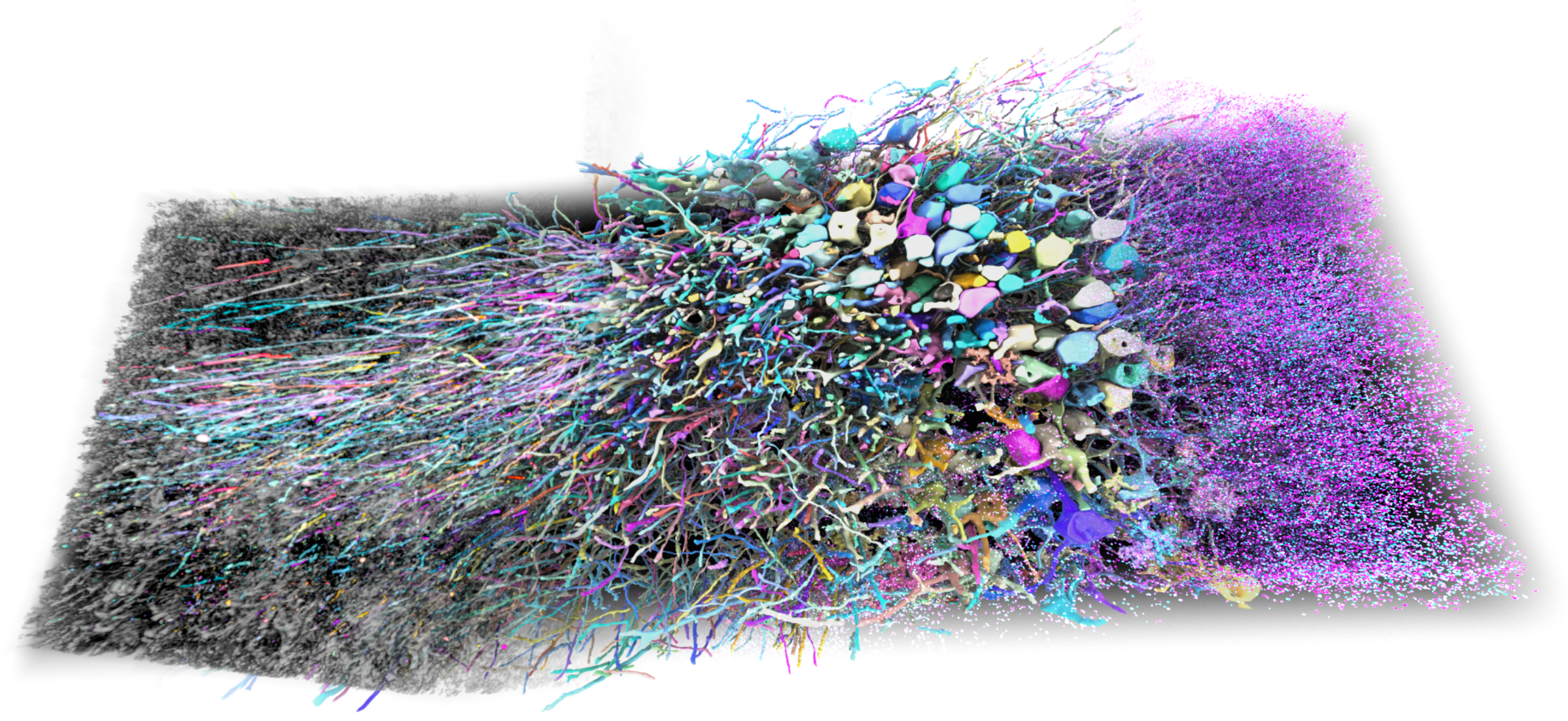
Protein-barcode Reconstruction by Iterative Staining with Molecular annotations (PRISM) is a set of methods and molecular tools enabling high-throughput reconstruction of individual neurons in brain tissue. PRISM begins with the delivery of stochastic combinations of cell-filling antigenically distinct protein tags (or "bits"), creating over 100,000 unique combinations detectable by antibodies. All of the PRISM protein tags have been validated for use in expansion microscopy, allowing for super-resolution light microscopy. In brain tissue, these protein barcodes allow for efficient machine-learning segmentation of neurons and proofreading of neuron reconstructions. This minimizes the need for human proofreading, the biggest cost driver for brain circuit mapping and connectomics.
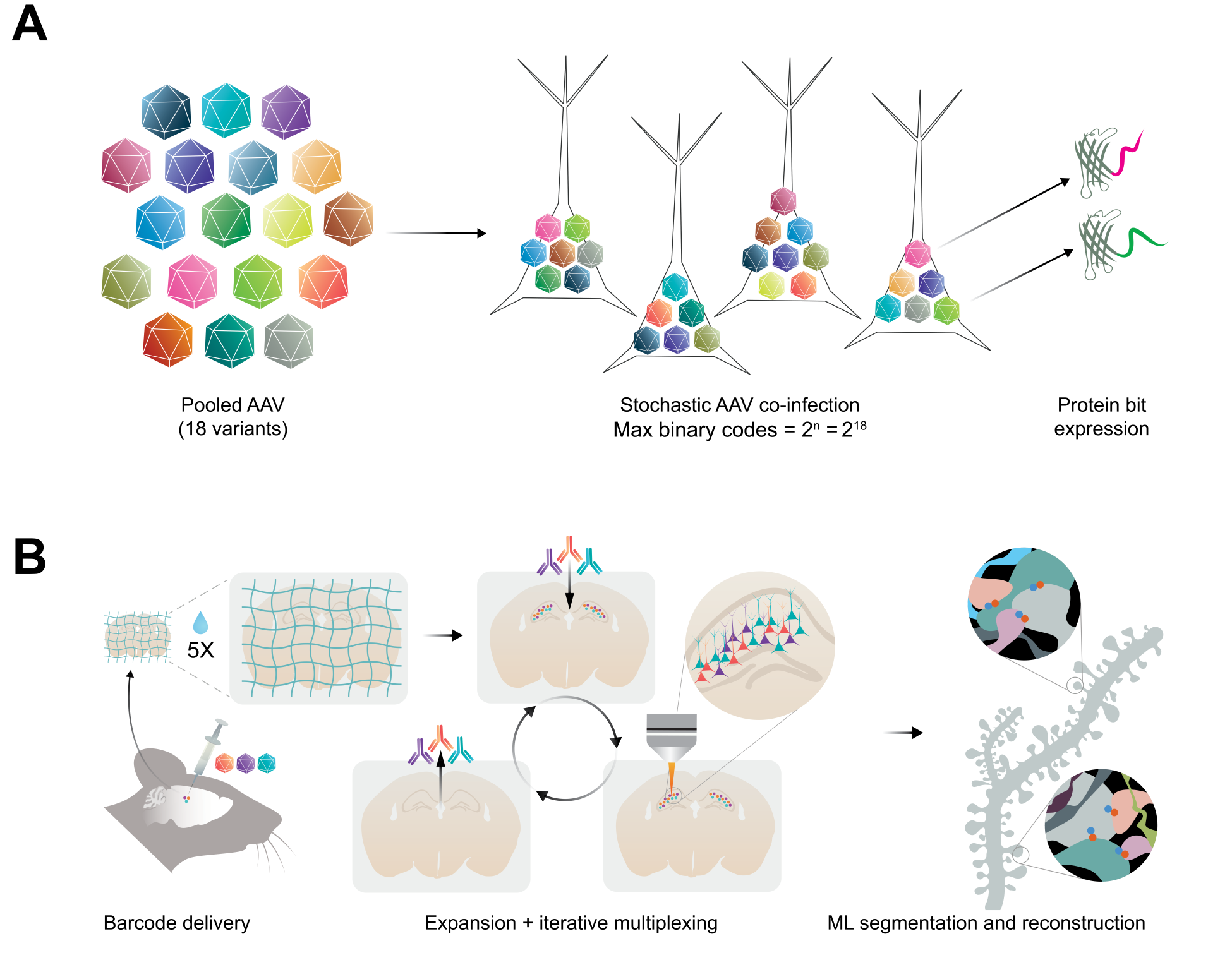
PRISM is part of an integrated pipeline, but individual components can be used separately for additional applications. Park & Sheridan et al. (2025) use plasmids expressing the barcode proteins under the constitutive CAG promoter (Addgene #242764–242781), but E11 Bio has also developed a set of Cre-dependent, hSyn-Tet inducible plasmids (Addgene #242782–242799) suitable for targeting specific neuron subtypes in Cre-driver lines. These could also be used in conjunction with Cre-enhancer viruses for highly focused circuit mapping and reconstruction. Potential applications could also include using individual or subsets of barcode proteins to label and detect multiple injection sites in the same tissue, as well as barcoding with specific enhancers for morphological readout.
How to Use PRISM
Barcodes are delivered as pooled AAVs either directly to a region of interest or systemically using a capsid capable of crossing the blood-brain barrier such as PHP.eB. The plasmids can be packaged as AAVs and pooled together for co-delivery. Alternatively, plasmids can be combined at equimolar ratios and packaged together in one AAV preparation. The depositing lab recommends keeping these production runs small to limit the amount of bias in packaging representation. To read out the barcodes, each protein tag can be imaged with a corresponding primary and dye-conjugated secondary antibody (as listed in Park & Sheridan et al., 2025). These reagents have been used in expansion microscopy workflows to achieve high-resolution optical imaging down to individual synapses.
PRISM Plasmids
| ID | Plasmid | Description | Tags |
|---|
Additional Resources
PRISM was developed by E11 Bio (Link opens in a new window), a non-profit focused research organization supported by Convergent Research (Link opens in a new window), in collaboration with The Francis Crick Institute (Link opens in a new window), MIT (Link opens in a new window), and The Max Planck Institute for Biological Intelligence (Link opens in a new window). Read more from E11 Bio (Link opens in a new window).
For more information about PRISM, see the preprint:
Park, S.Y., Sheridan, A., An, B., Jarvis, E., Lyudchik, J., Patton, W., Axup, J.Y., Chan, S.W., Damstra, H.G.J., Leible, D., Leung, K.S., Magno, C.A., Meeran, A., Michalska, J.M., Rieger, F., Wang, C., Wu, M., Church, G.M., ... Rodriques, S.G., & Payne, A.C. (2025). Combinatorial protein barcodes enable self-correcting neuron tracing with nanoscale molecular context. bioRxiv 2025.09.26.678648; doi: https://doi.org/10.1101/2025.09.26.678648 (Link opens in a new window)
More Resources at Addgene:
Content last reviewed: 5 September 2025


