We narrowed to 17,255 results for: form
-
Plasmid#128372PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only
-
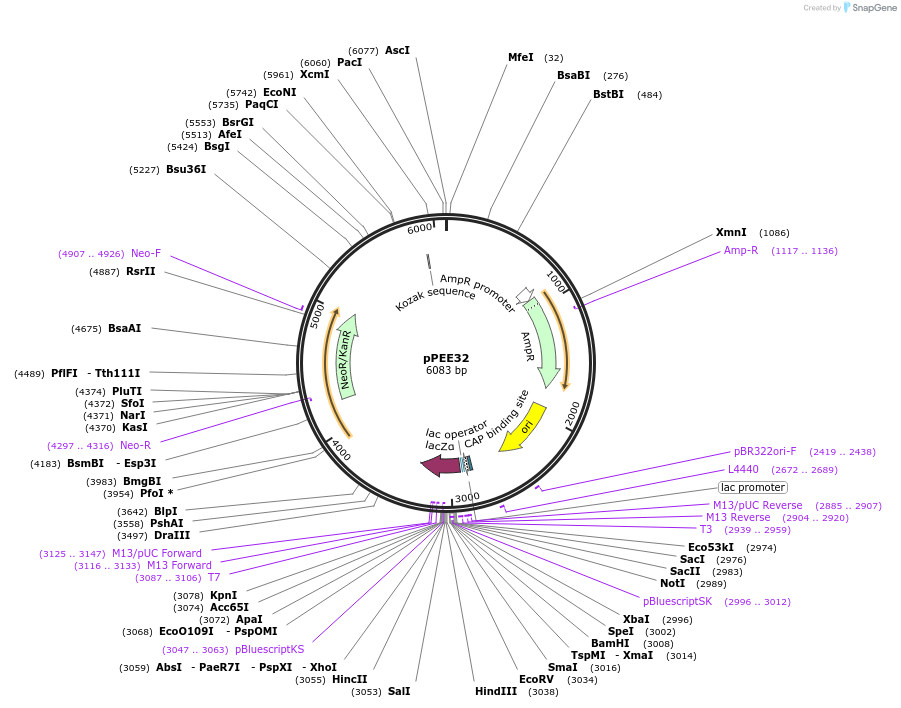
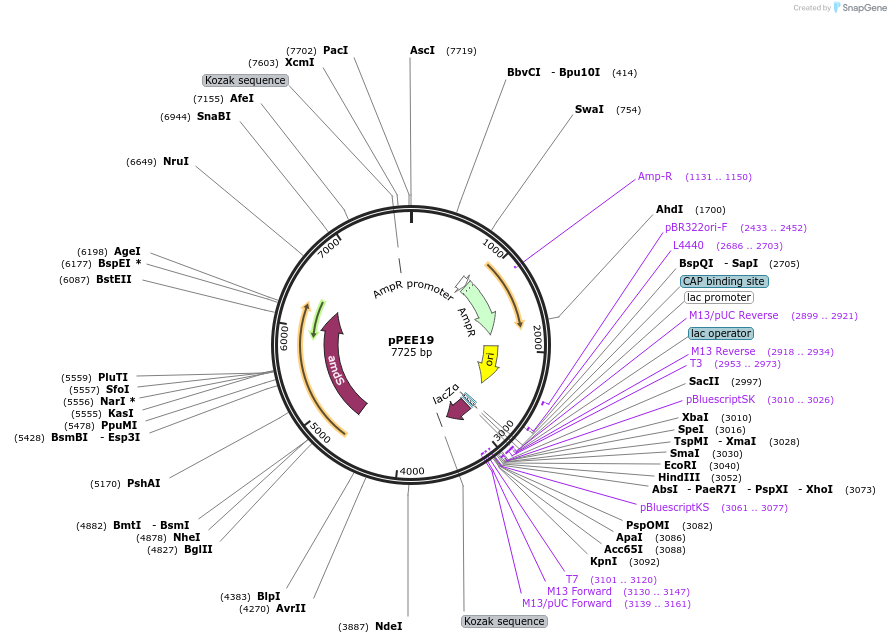
pPEE20
Plasmid#128363PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceOct. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
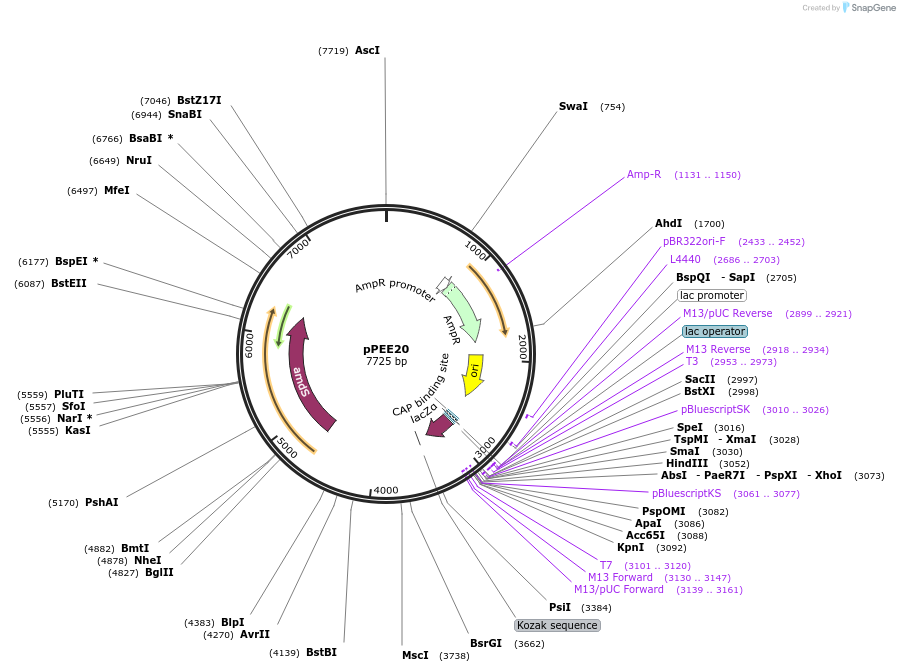
pPEE35
Plasmid#128375PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
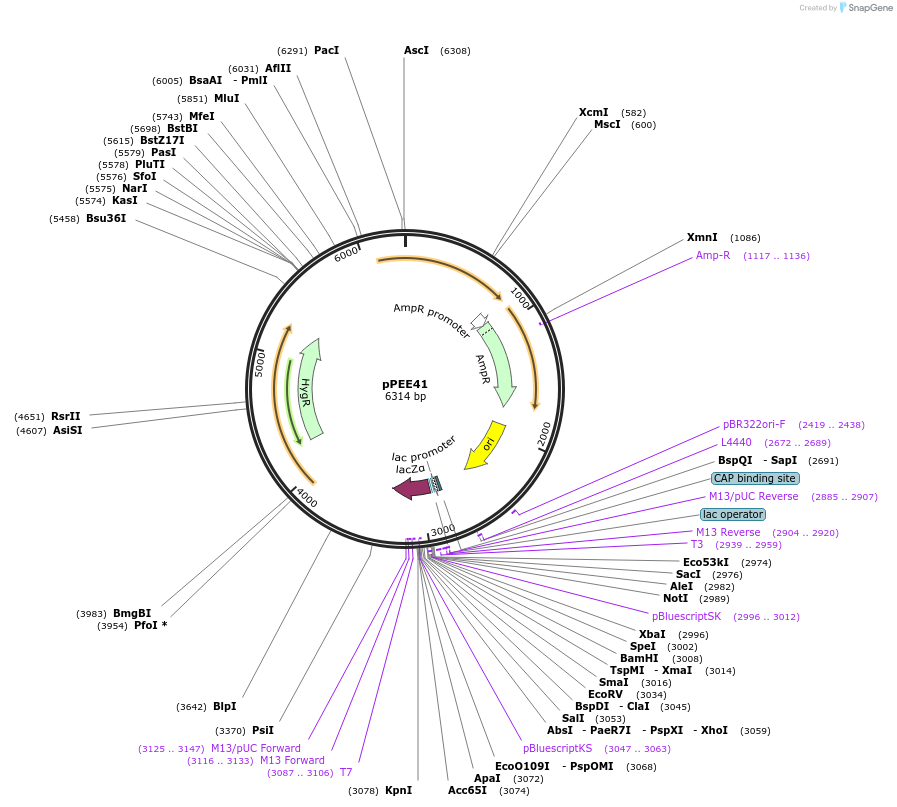
pPEE41
Plasmid#128381PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
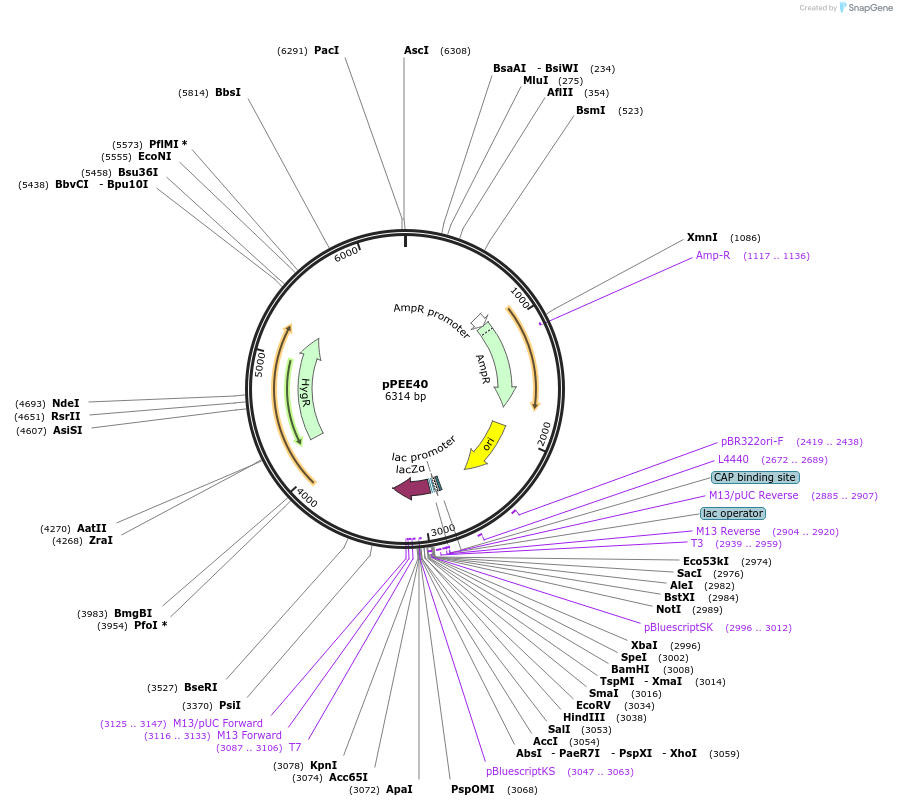
pPEE40
Plasmid#128380PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
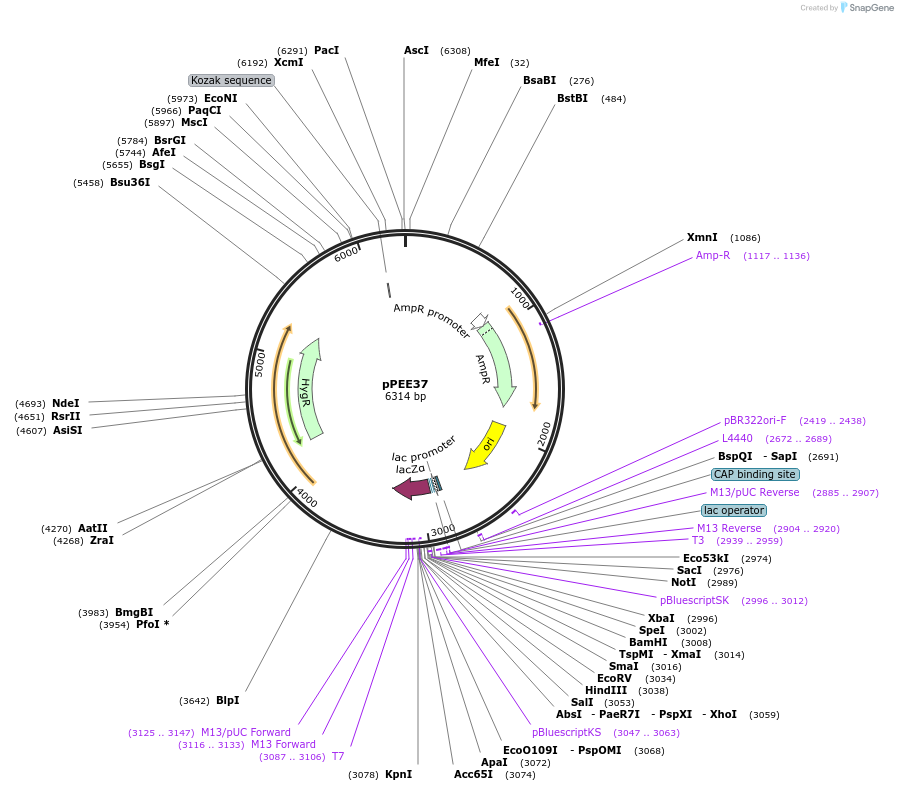
pPEE37
Plasmid#128377PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
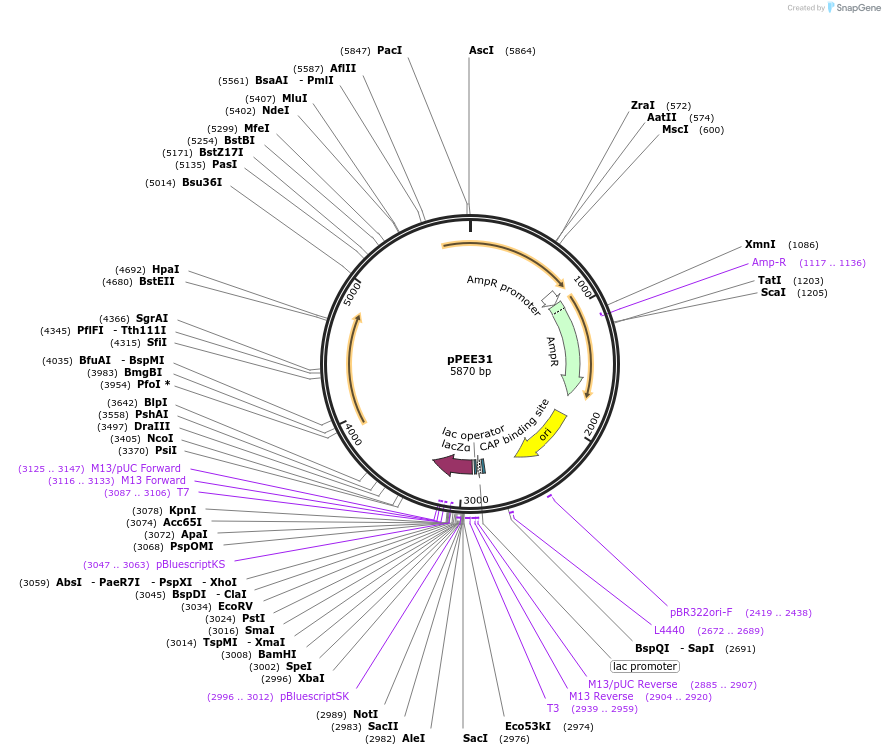
pPEE31
Plasmid#128371PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
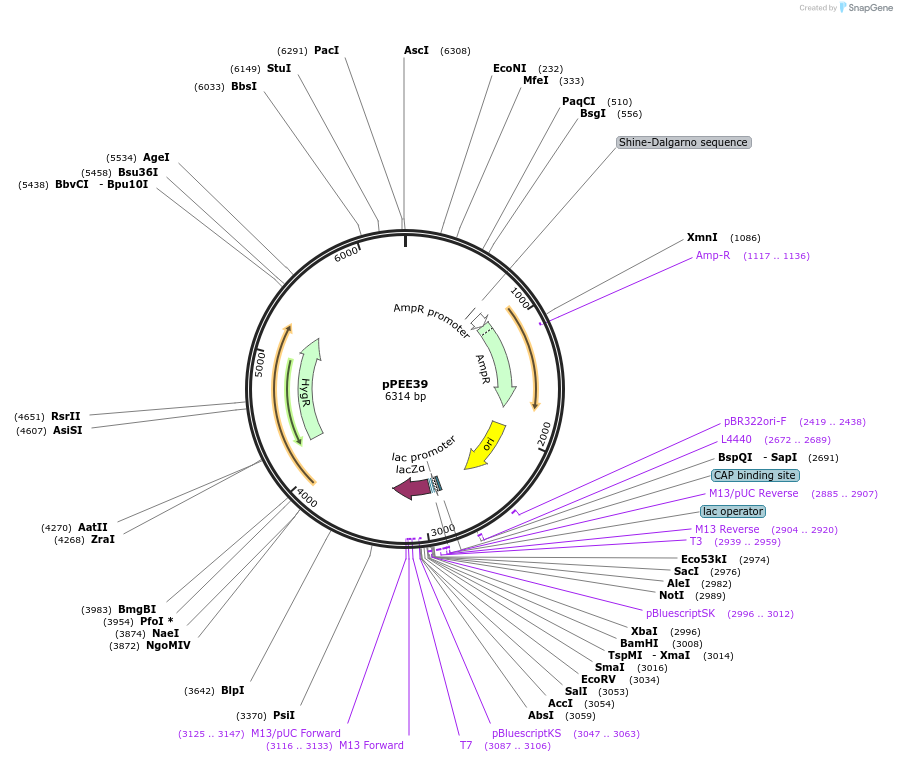
pPEE39
Plasmid#128379PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
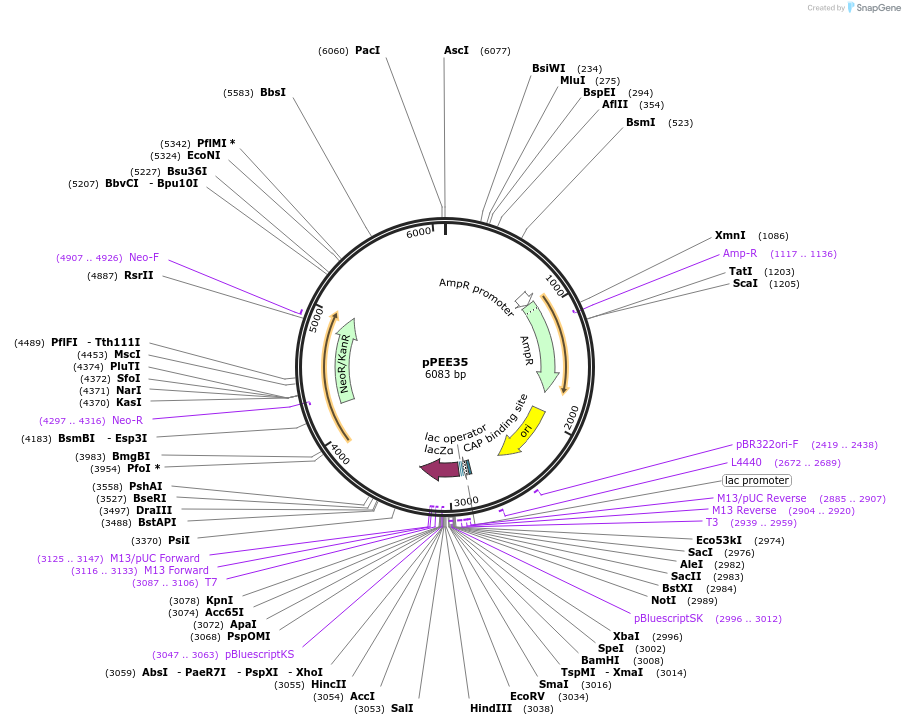
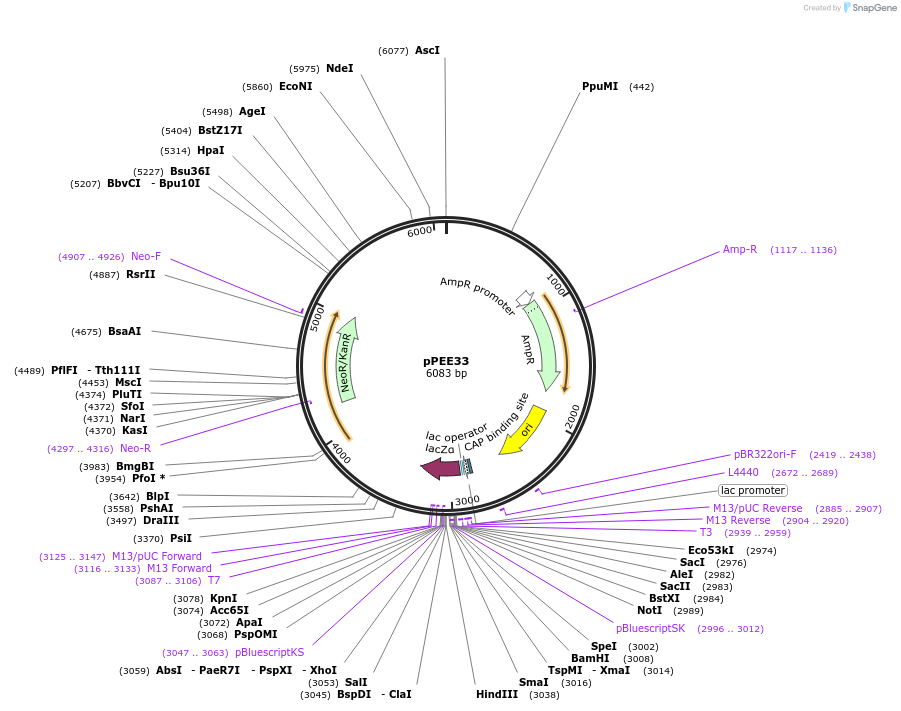
pPEE33
Plasmid#128373PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
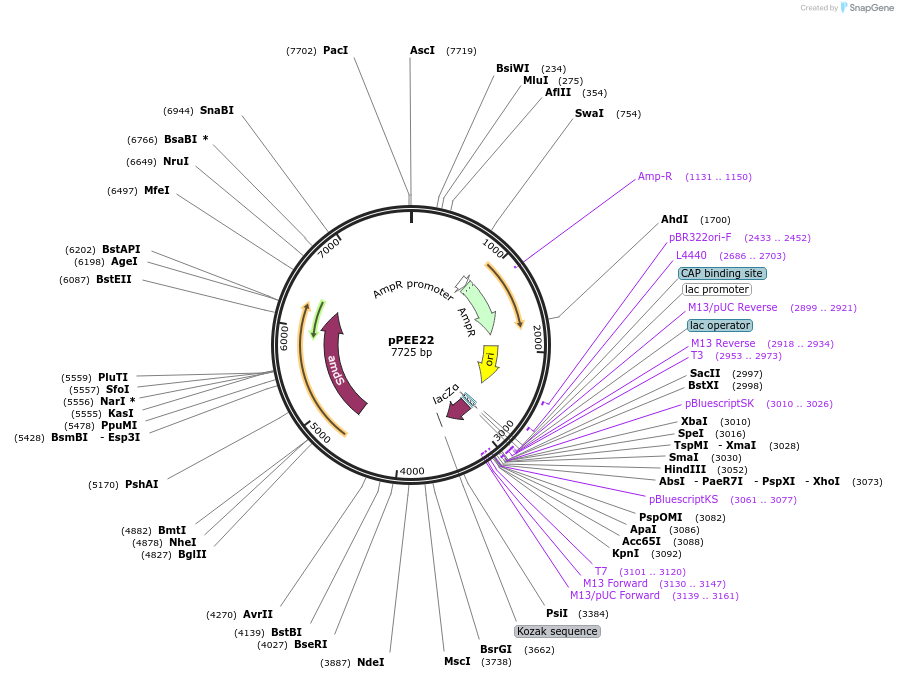
pPEE22
Plasmid#128365PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
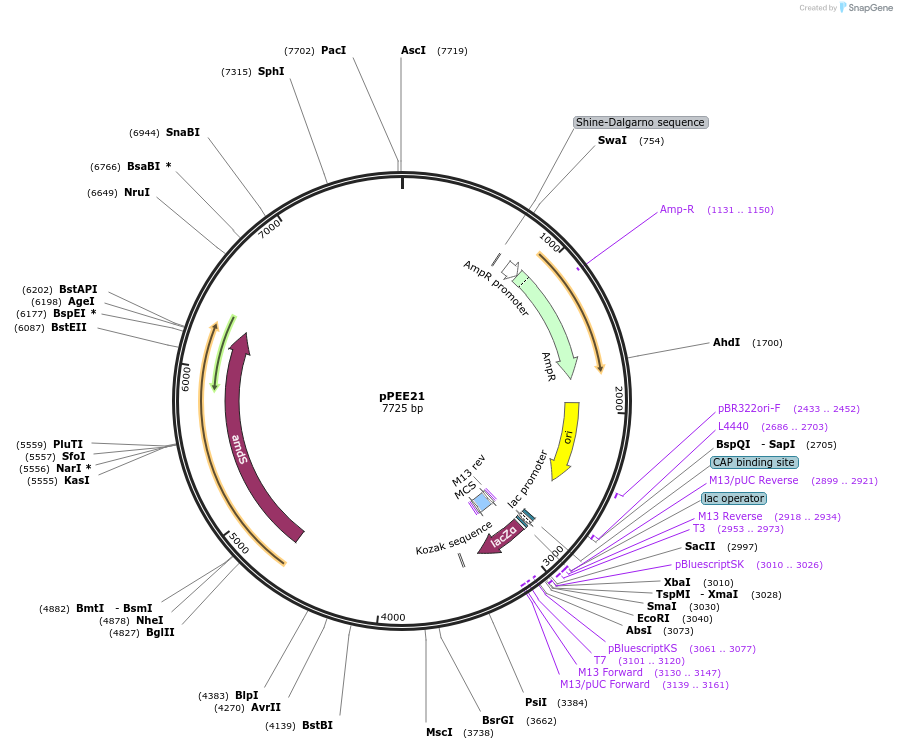
pPEE21
Plasmid#128364PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
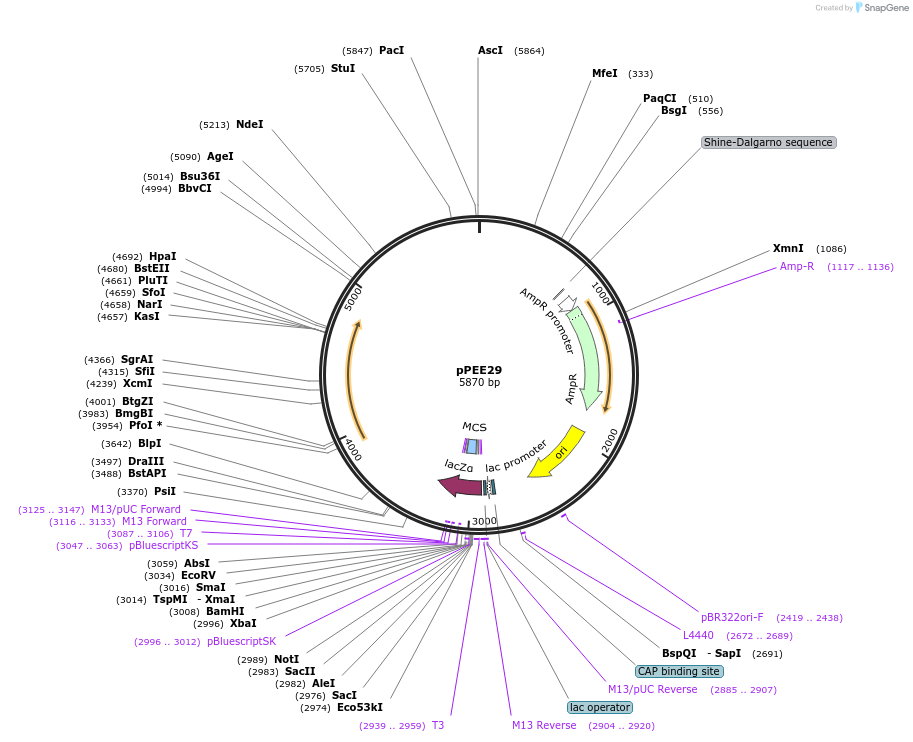
pPEE29
Plasmid#128369PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
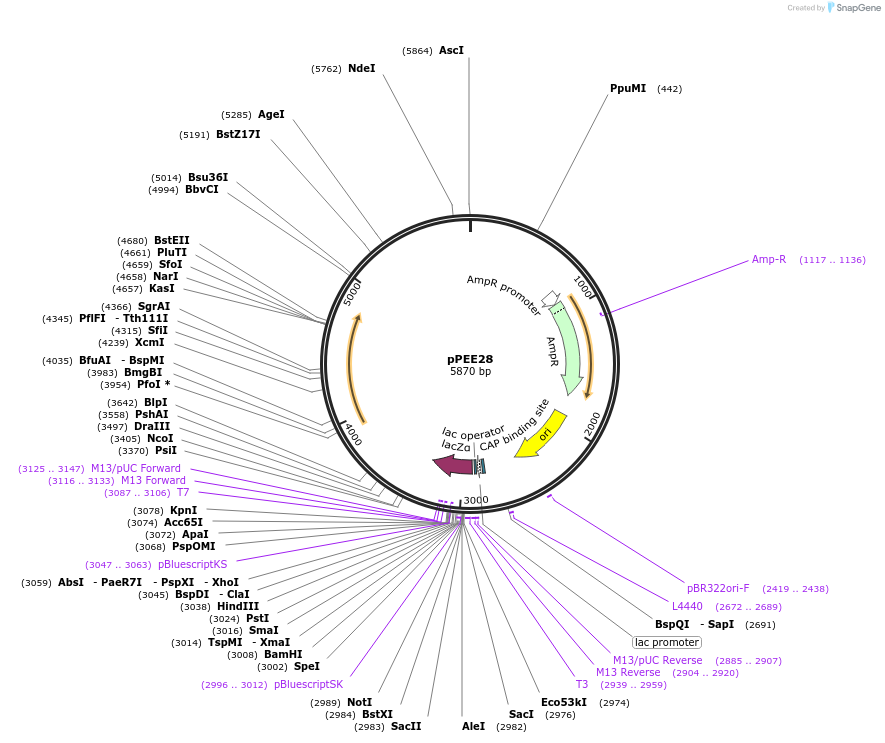
pPEE28
Plasmid#128368PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
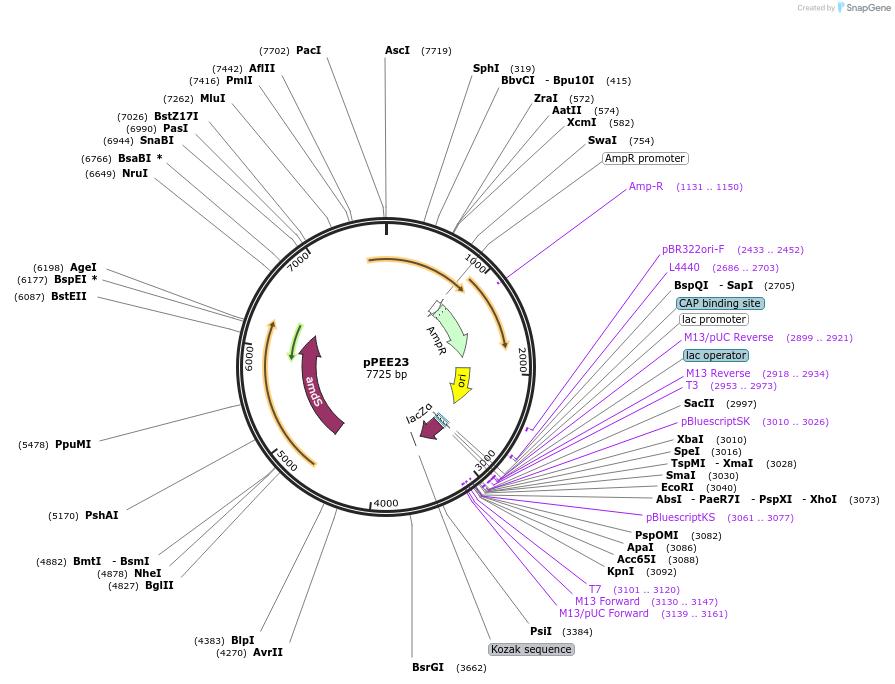
pPEE23
Plasmid#128366PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
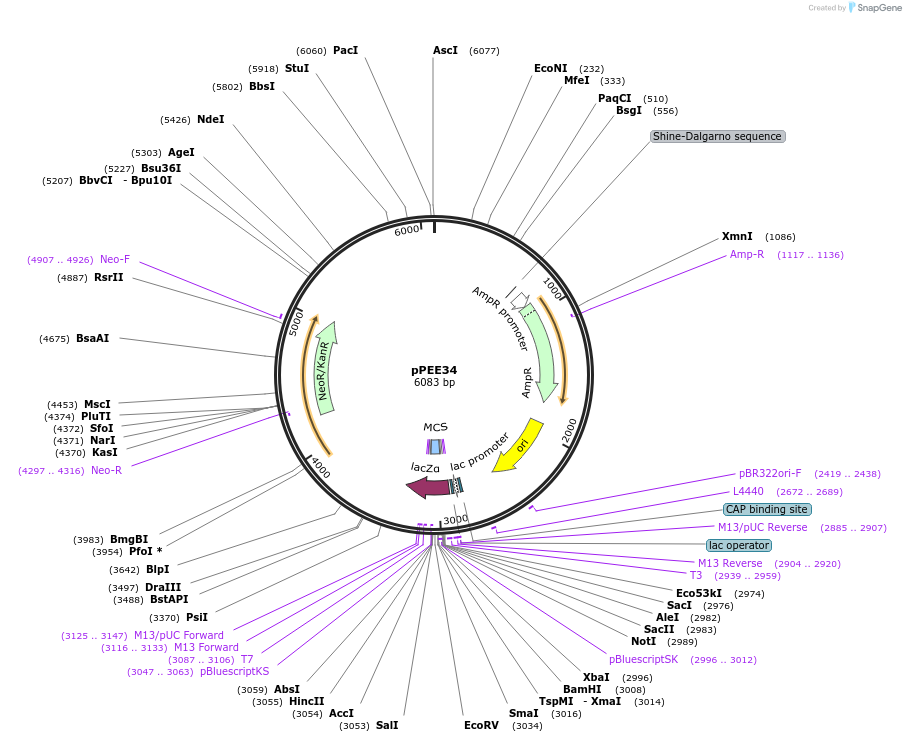
pPEE34
Plasmid#128374PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
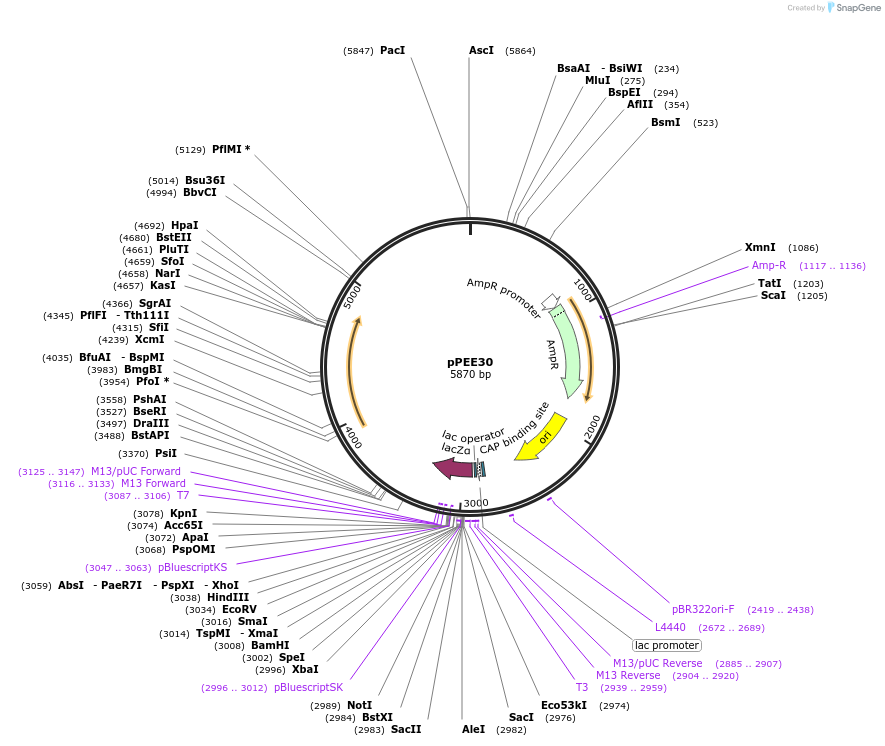
pPEE30
Plasmid#128370PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
pPEE19
Plasmid#128362PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
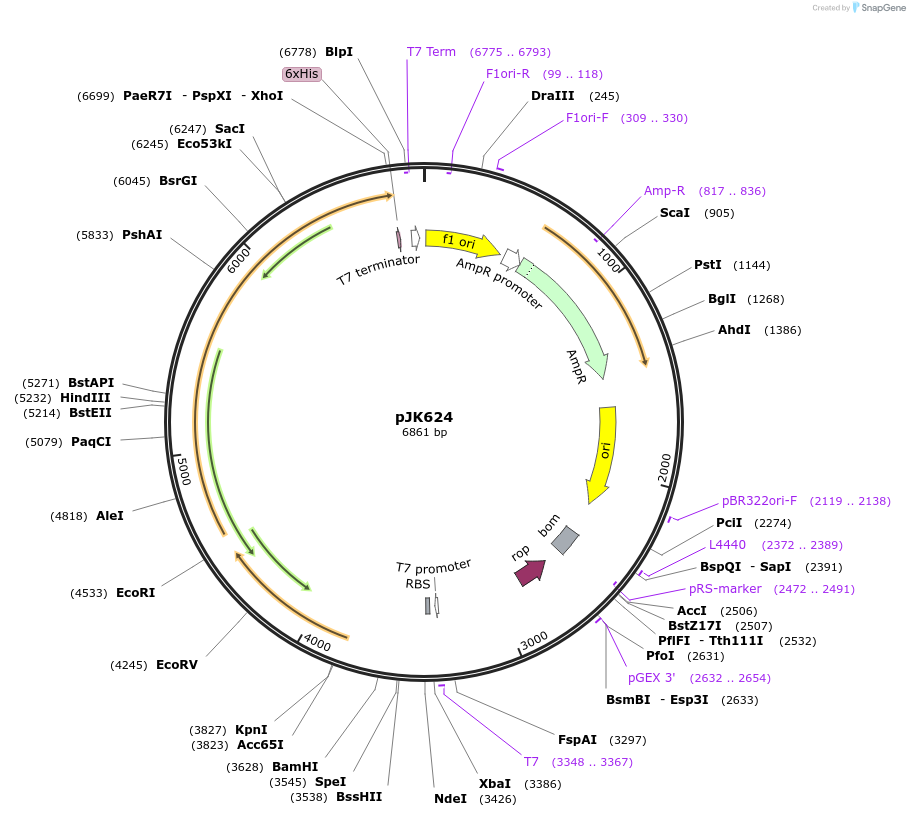
pJK624
Plasmid#72539PurposeProduces Acetobacter aceti 1023 OrfY; formylglycinamidine ribonucleotide synthetase glutaminase subunit; large subunit with C-terminal His6 tag (AaOrfY, AaPurQ, AaPurLH6)DepositorInsertshypothetical protein in purSQL operon
formylglycinamidine ribonucleotide synthetase, glutaminase subunit
formylglycinamidine ribonucleotide synthetase, large subunit
TagsHis6ExpressionBacterialMutationsynthetic genePromoterT7 and n/aAvailable SinceJan. 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
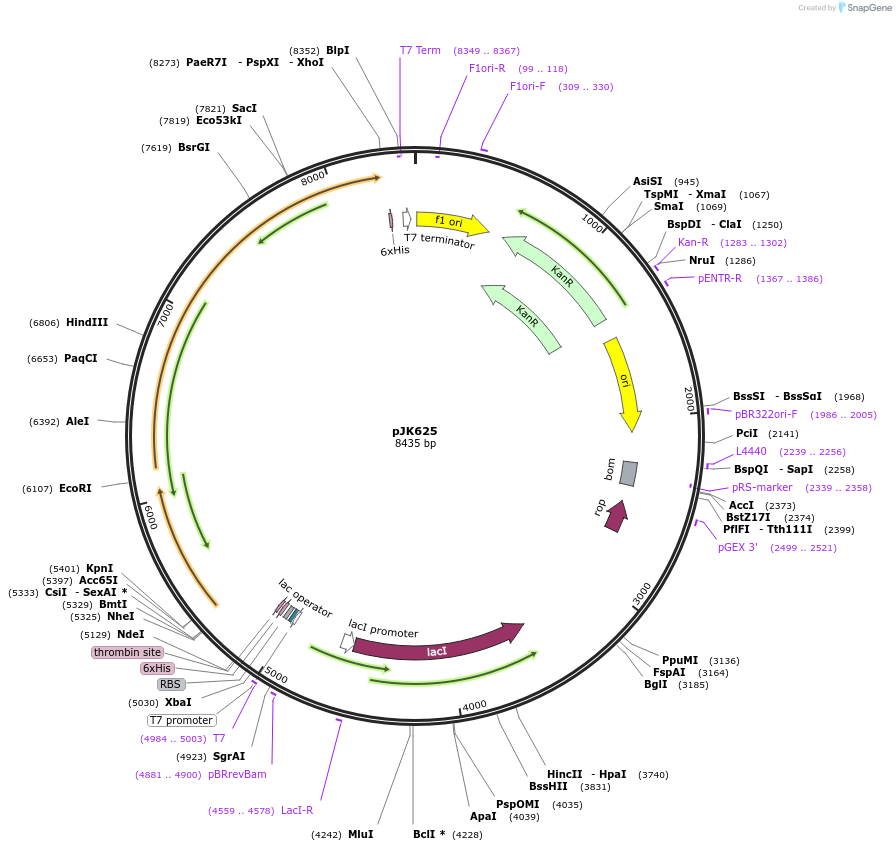
pJK625
Plasmid#72541PurposeProduces Acetobacter aceti 1023 formylglycinamidine ribonucleotide synthetase small subunit with N-terminal His6 tag (H6AaPurS); glutaminase subunit (AaPurQ); large subunit (AaPurL)DepositorInsertshypothetical protein in purSQL operon
formylglycinamidine ribonucleotide synthetase, glutaminase subunit
formylglycinamidine ribonucleotide synthetase, large subunit
TagsHis6ExpressionBacterialMutationsynthetic genePromoterT7 and n/aAvailable SinceJan. 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
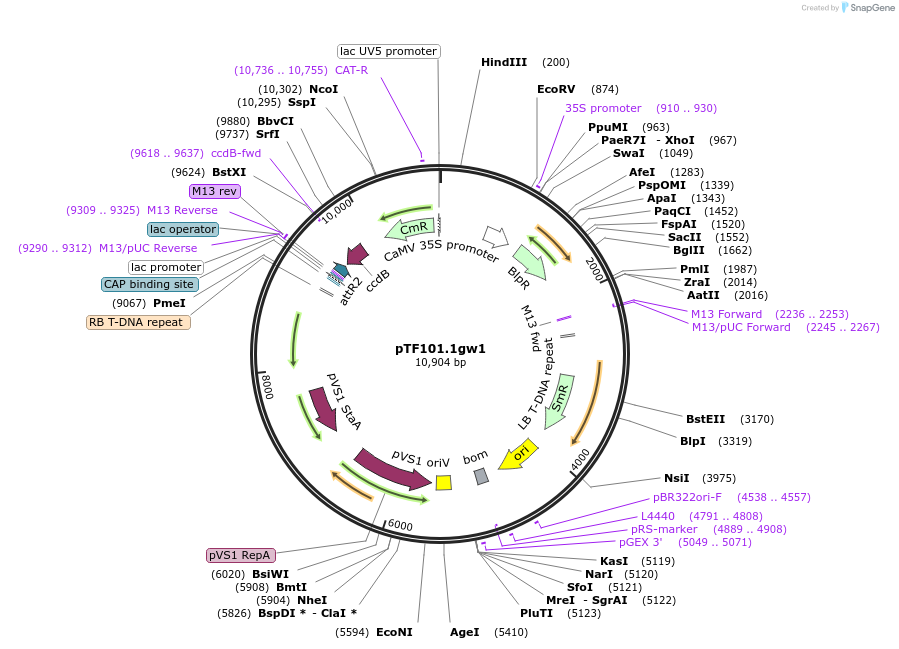
pTF101.1gw1
Plasmid#134768PurposeA binary plasmid for gateway 1-Entry in pTF101.1 background (attR1 & attR2) containing double CaMV 35S promoter and TEV enhancer to drive bialaphos resistance gene (bar) for plant transformationDepositorArticleTypeEmpty backboneExpressionPlantAvailable SinceDec. 10, 2019AvailabilityAcademic Institutions and Nonprofits only