We narrowed to 1,735 results for: nif
-
Plasmid#165515PurposeMethylase recognizes double-stranded sequence GAATTC, methylates A-3 on both strands, and protects DNA from cleavage by EcoRI.DepositorInsertM.EcoRI
UseSynthetic BiologyAvailable SinceJuly 29, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
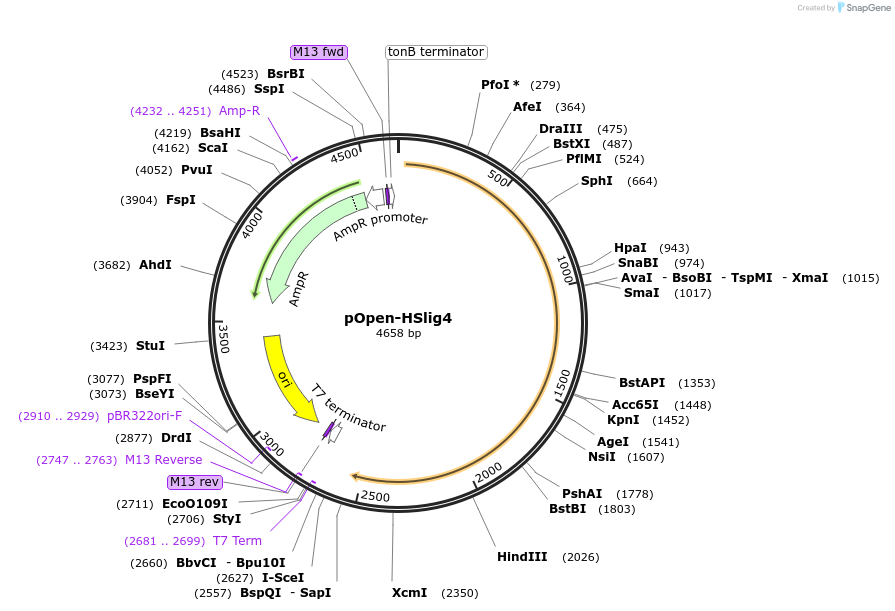
pOpen-HSlig4
Plasmid#165503PurposeLigase used in efficiently joining single-strand breaks in a double-strand polydeoxynucleotide in an ATP-dependent reaction.DepositorInsertDNA Ligase IV
UseSynthetic BiologyAvailable SinceJuly 27, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
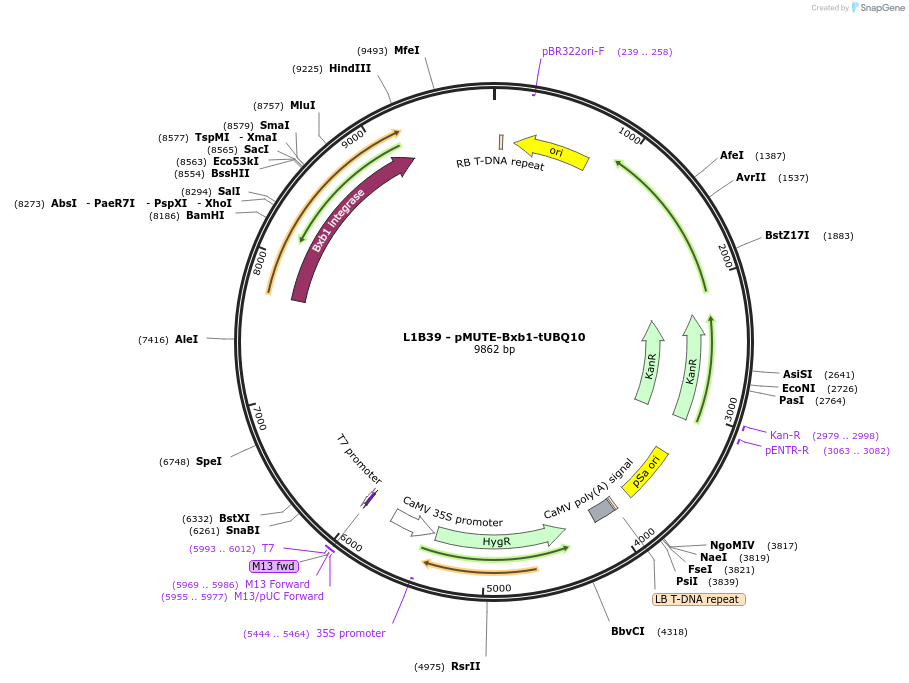
L1B39 - pMUTE-Bxb1-tUBQ10
Plasmid#219704PurposeLevel 1 pMUTE-Bxb1-tUBQ10, for stomata specific expression of Bxb1 integraseDepositorInsertpMUTE:Bxb1:tUBQ10
ExpressionPlantAvailable SinceOct. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
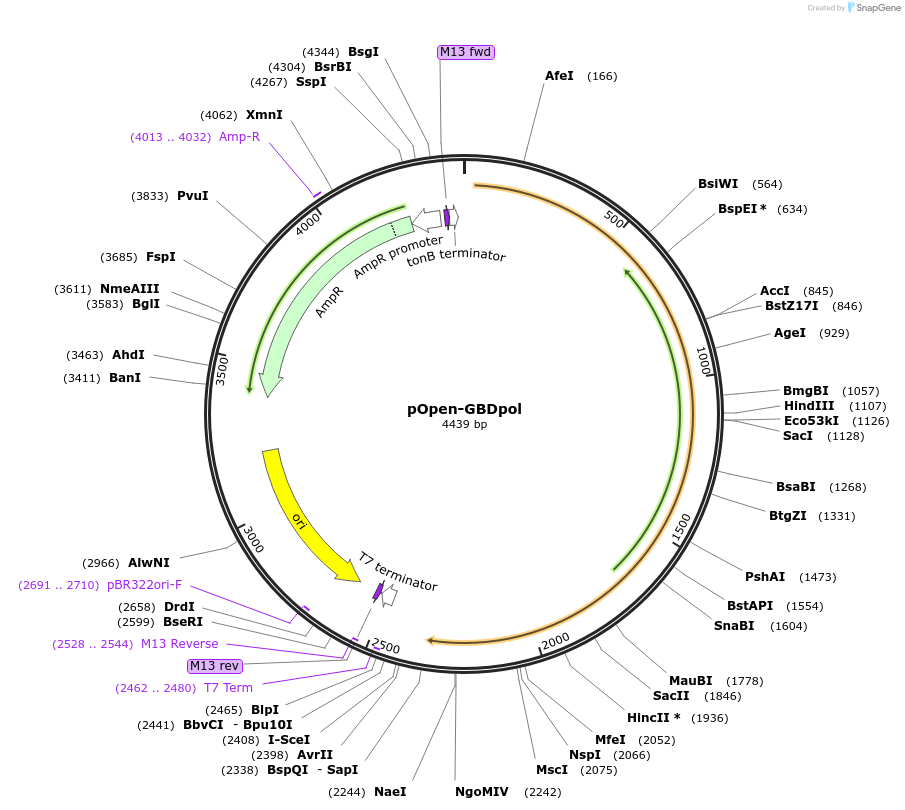
pOpen-GBDpol
Plasmid#165501PurposeRobust and extremely thermostable polymerase with 5x higher fidelity than Taq. Ideal for GC-rich or looped sequences.DepositorInsertPyrococcus Sp. Heat-Stable DNA Polymerase
UseSynthetic BiologyAvailable SinceJuly 26, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
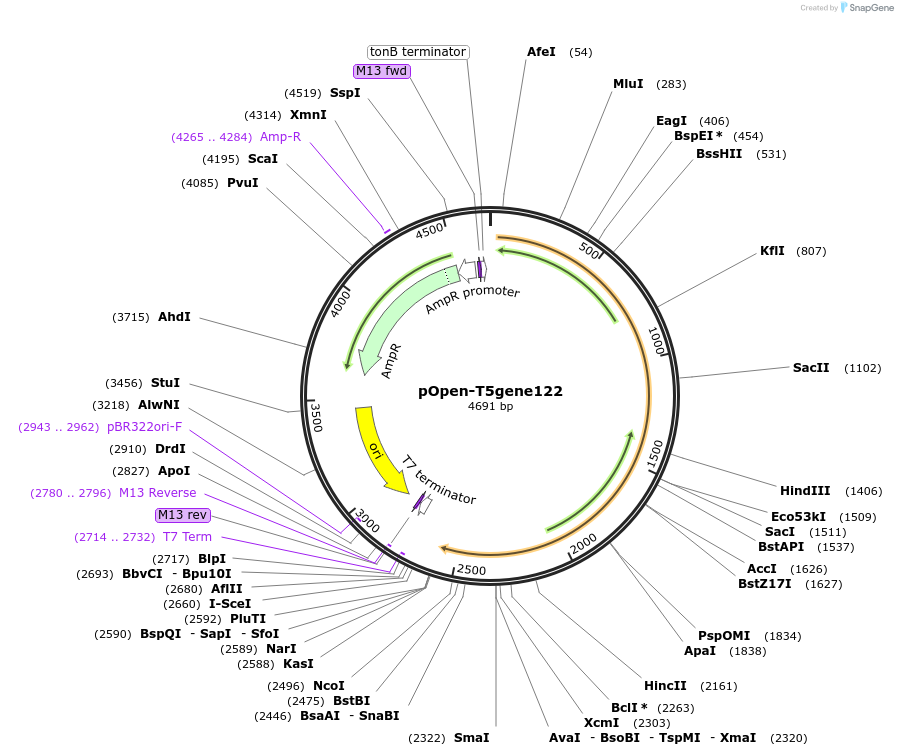
pOpen-T5gene122
Plasmid#165544PurposePossesses two enzymatic activities: DNA synthesis (polymerase) and exonucleolytic activity that degrades ssDNA in the 3'-5' direction for proofreading purposes.DepositorInsertT5 DNA Polymerase
UseSynthetic BiologyAvailable SinceAug. 9, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
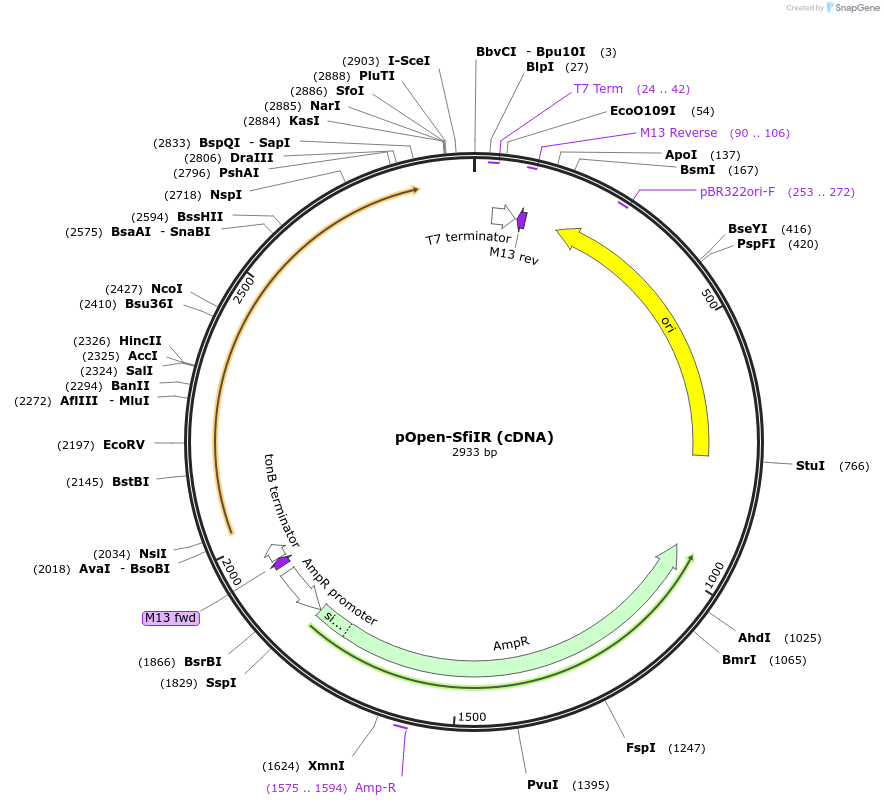
pOpen-SfiIR (cDNA)
Plasmid#165528PurposeType II restriction enzyme that recognizes the double-stranded sequence GGCCNNNNNGGCC and cleaves after N-4.DepositorInsertSfiI (used to create cDNA libraries)
UseSynthetic BiologyAvailable SinceAug. 5, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
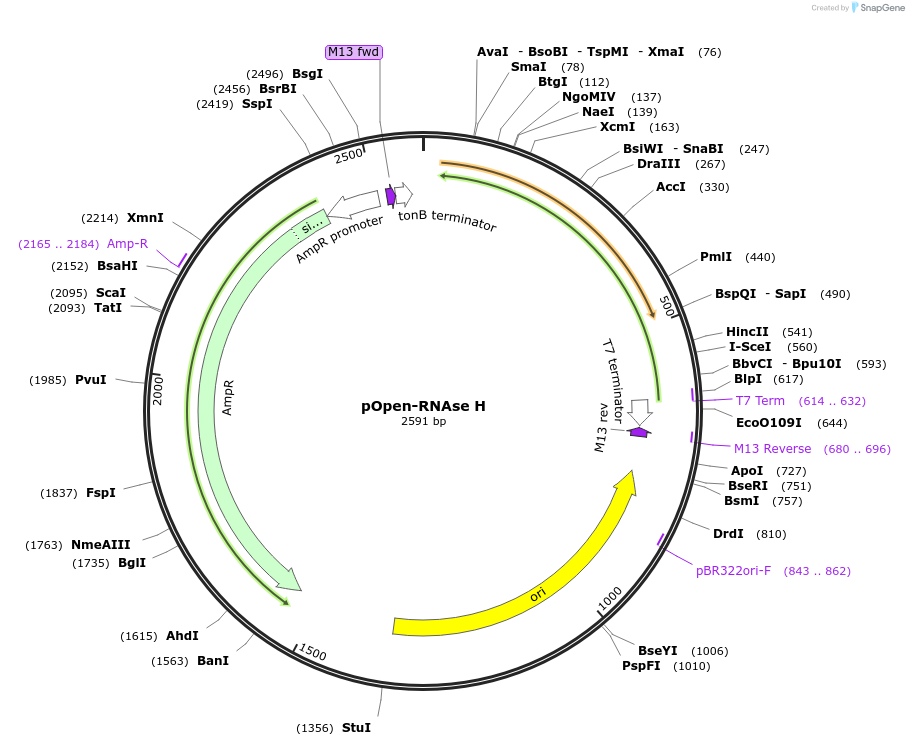
pOpen-RNAse H
Plasmid#165575PurposeEndonuclease that specifically degrades the RNA of RNA-DNA hybrids. Participates in DNA replication; helps to specify the origin of genomic replication.DepositorInsertRNAse H
UseSynthetic BiologyAvailable SinceAug. 24, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
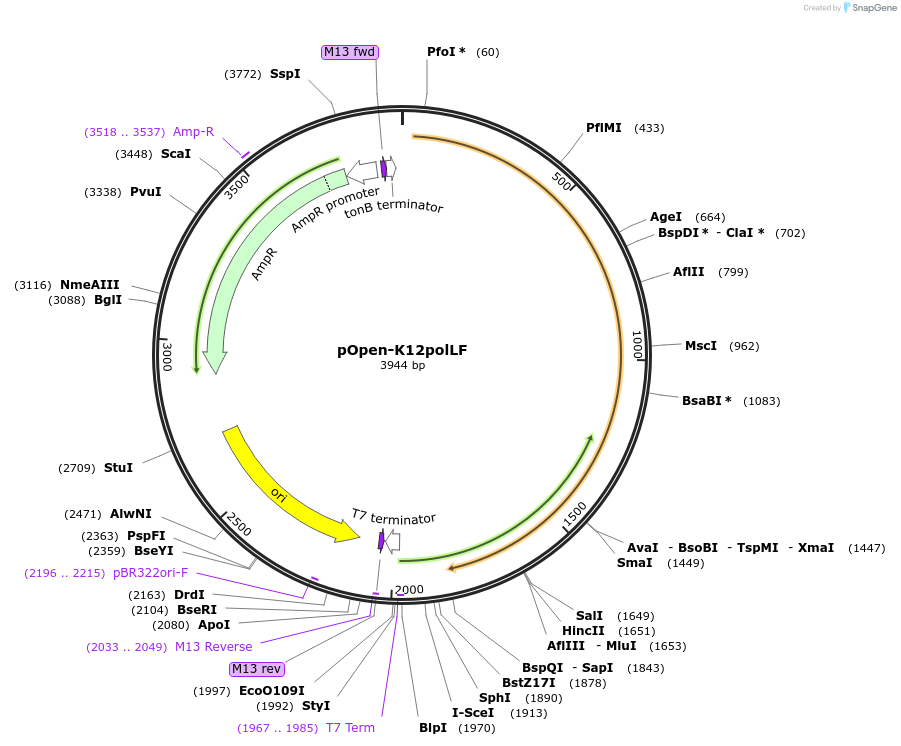
pOpen-K12polLF
Plasmid#165511PurposeDNA pol fragment that retains 5'-3' polymerase activity and 3'-5' exonuclease activity but loses 5'-3' exonuclease activity.DepositorInsertDNA Polymerase I, Large (Klenow) Fragment
UseSynthetic BiologyAvailable SinceJuly 30, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
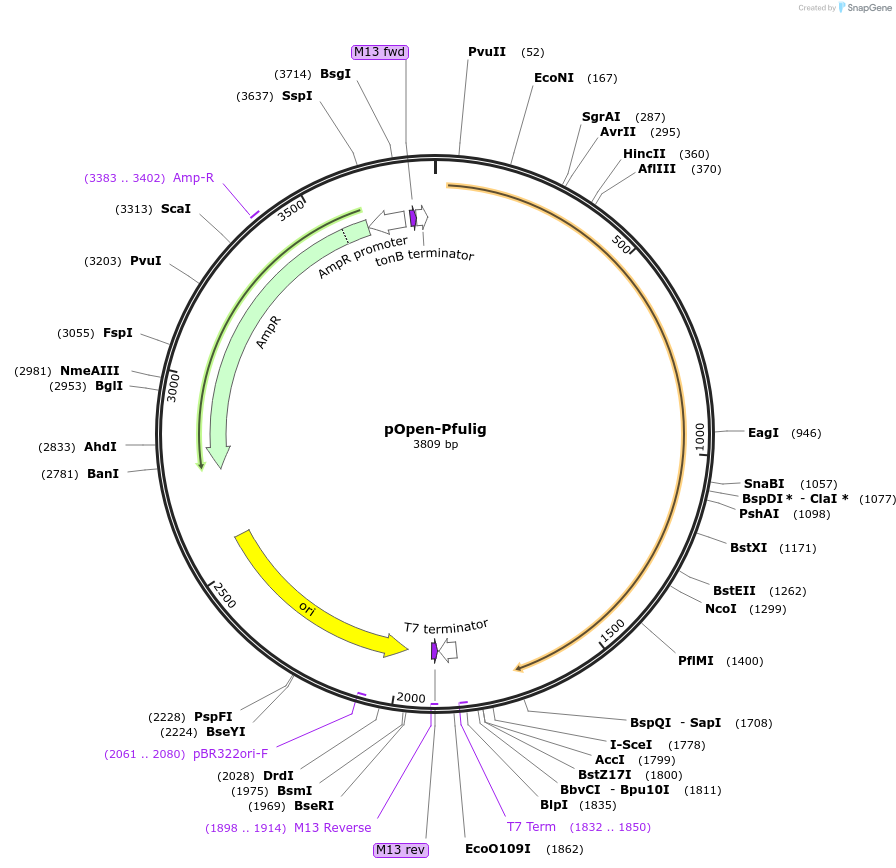
pOpen-Pfulig
Plasmid#165525PurposeDNA ligase that seals nicks in double-stranded DNA during DNA replication, DNA recombination and DNA repair.DepositorInsertPfu DNA Ligase
UseSynthetic BiologyAvailable SinceAug. 4, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
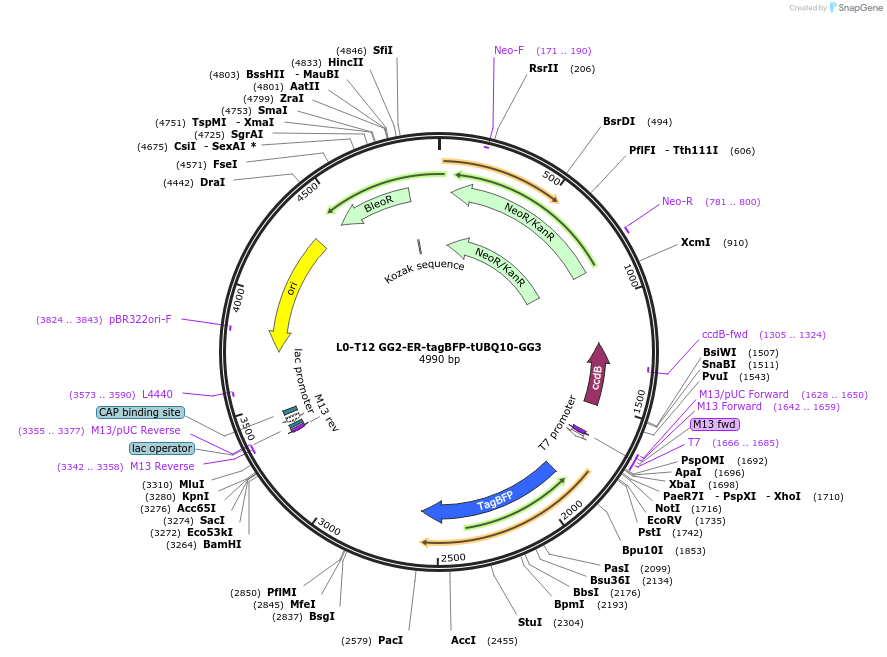
L0-T12 GG2-ER-tagBFP-tUBQ10-GG3
Plasmid#219695PurposeLevel 0 to construct history-dependent targetDepositorInsertGG2-ER-tagBFP-tUBQ10-GG3
UseSynthetic BiologyAvailable SinceJuly 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
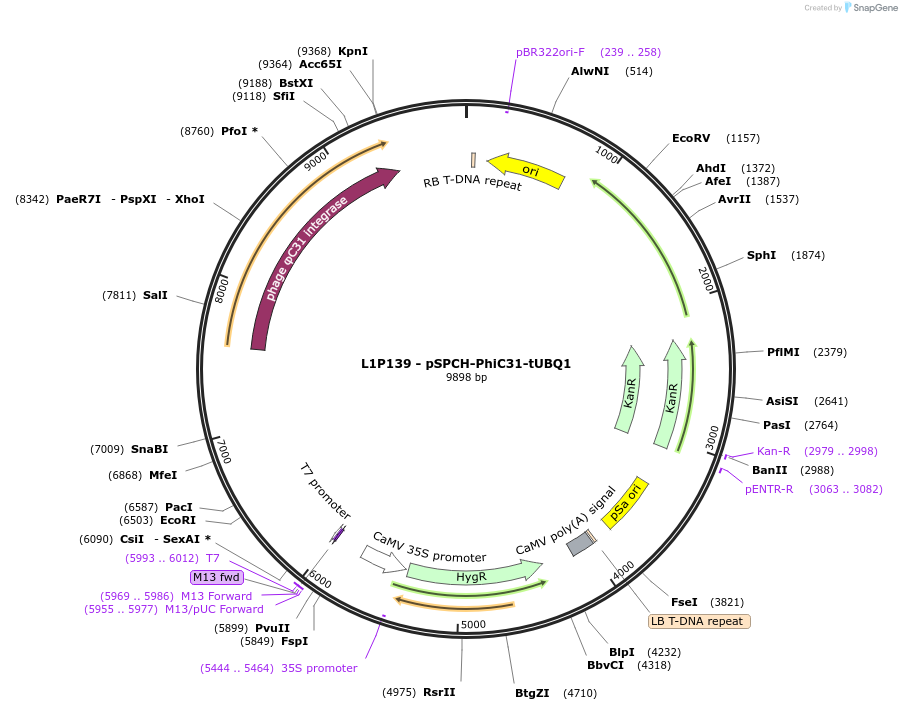
L1P139 - pSPCH-PhiC31-tUBQ1
Plasmid#219703PurposeLevel 1 pSPCH-PhiC31-tUBQ1, for early stage of stomata development expression of PhiC31 integraseDepositorInsertpSPCH:PhiC31:tUBQ1
ExpressionPlantAvailable SinceJune 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
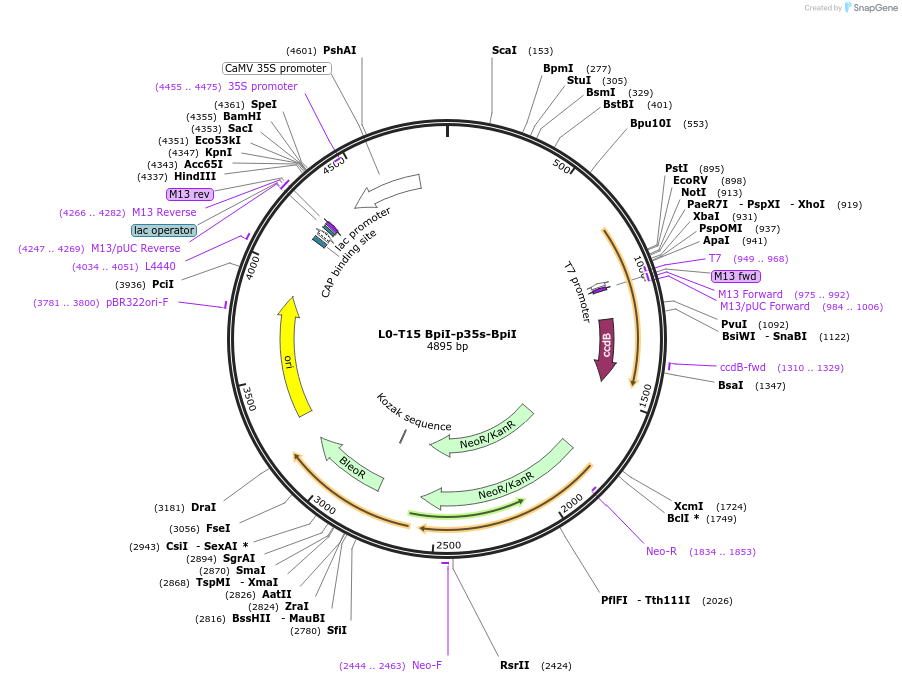
L0-T15 BpiI-p35s-BpiI
Plasmid#219698PurposeLevel 0 to insert p35S promoter to history-dependent targetDepositorInsertBpiI-p35s-BpiI
UseSynthetic BiologyAvailable SinceJune 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
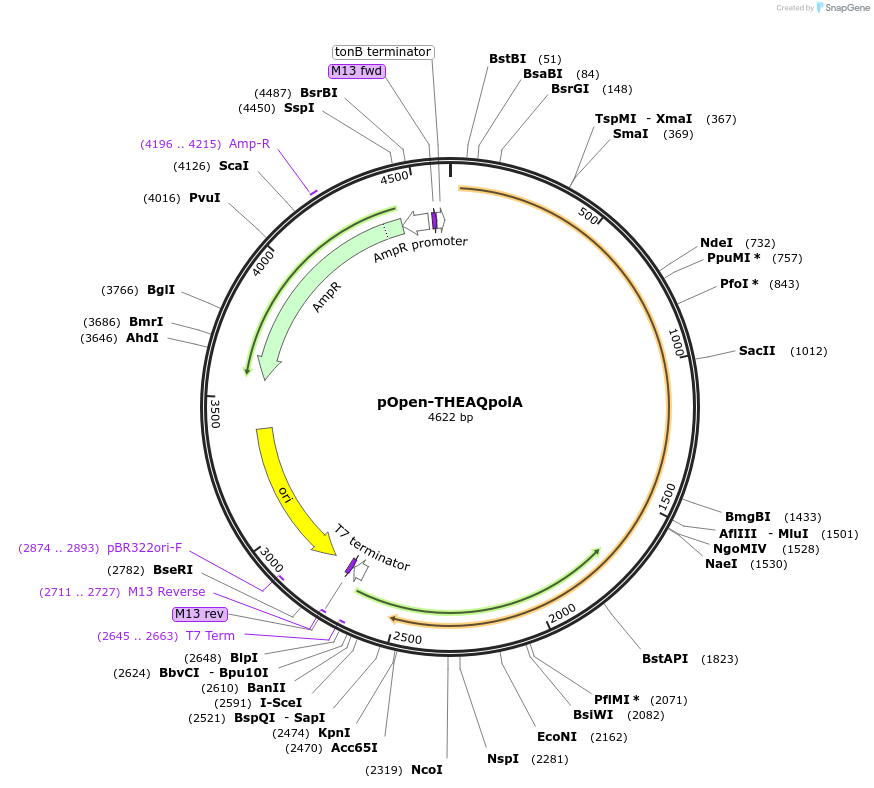
pOpen-THEAQpolA
Plasmid#165512PurposeDNA polymerase used in routine PCR method because of its high thermostability, standard testing, screening and output testing.DepositorInsertTaq DNA Polymerase
UseSynthetic BiologyAvailable SinceAug. 9, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
pOpen-T7gene5
Plasmid#165534PurposeDNA Pol with high fidelity and rapid extension rate (useful in copying long stretches of DNA); Has strong 3'-5' exonuclease.DepositorInsertT7 DNA Polymerase (unmodified)
UseSynthetic BiologyAvailable SinceAug. 5, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
pOpen-T3gene1
Plasmid#165576PurposeDNA-dependent RNA polymerase derived from T3 bacteriophage that catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates.DepositorInsertT3 RNA Polymerase
UseSynthetic BiologyAvailable SinceAug. 24, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
pOpen-dbh
Plasmid#165513PurposeThermostable gamma-family lesion-bypass DNA Polymerase that efficiently synthesizes DNA across a variety of DNA template lesions.DepositorInsertSulfolobus DNA Polymerase IV
UseSynthetic BiologyAvailable SinceJuly 30, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
L1P137 - pAHP6-PhiC31-NLS-tUBQ1
Plasmid#219702PurposeLevel 1 pAHP6-PhiC31-NLS-tUBQ1, for xylem pole specific expression of PhiC31 integraseDepositorInsertpAHP6:PhiC31-NLS:tUBQ1
ExpressionPlantAvailable SinceJune 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
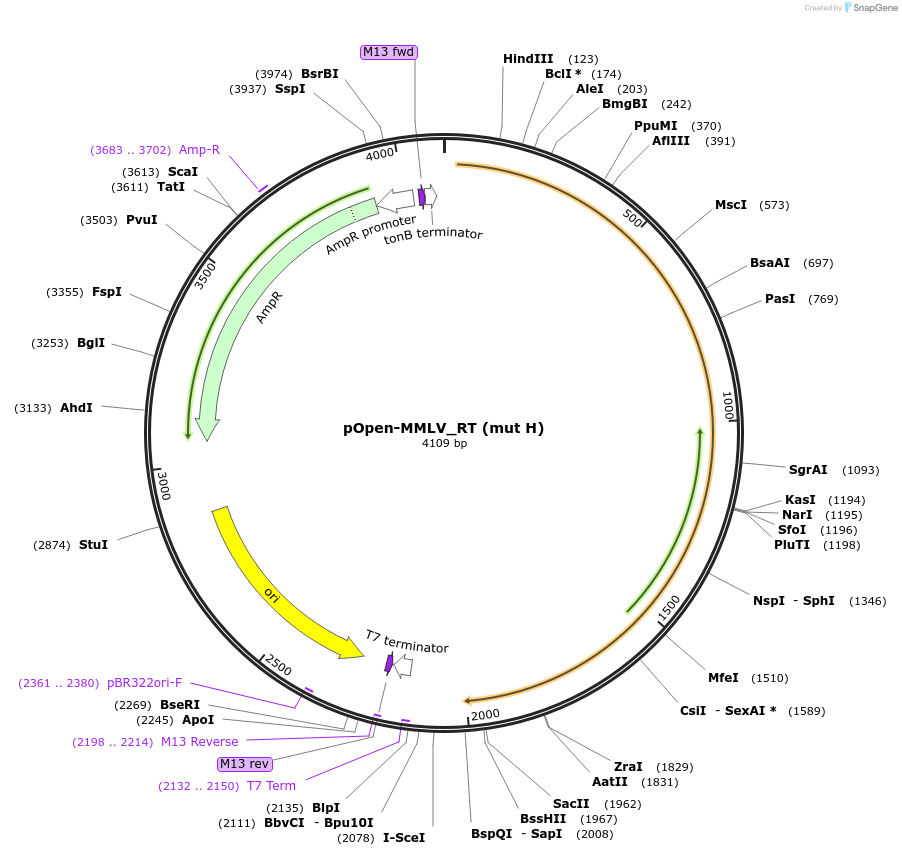
pOpen-MMLV_RT (mut H)
Plasmid#165546PurposeSingle 75 kDa monomer, cDNA synthesis; high enzyme activity and processivity.DepositorInsertMoloney Murine Leukemia Virus (MMLV) Reverse Transcriptase (RNAse H deactivated by 3 mutations)
UseSynthetic BiologyAvailable SinceAug. 10, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
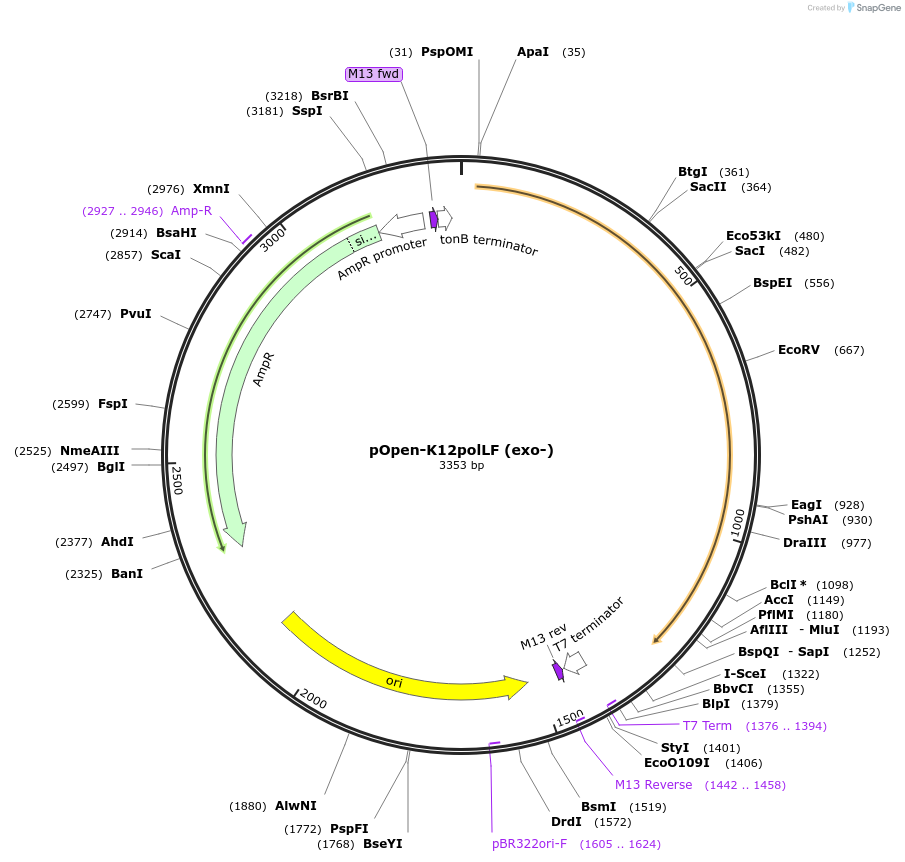
pOpen-K12polLF (exo-)
Plasmid#165522PurposeDNA pol fragment used in flourescent labelling for microarray, dA and dT tailing, and ligating DNA adapters to DNA fragments.DepositorInsertDNA Polymerase I, Large (Klenow) Fragment (3'-5' exo-)
UseSynthetic BiologyAvailable SinceDec. 21, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
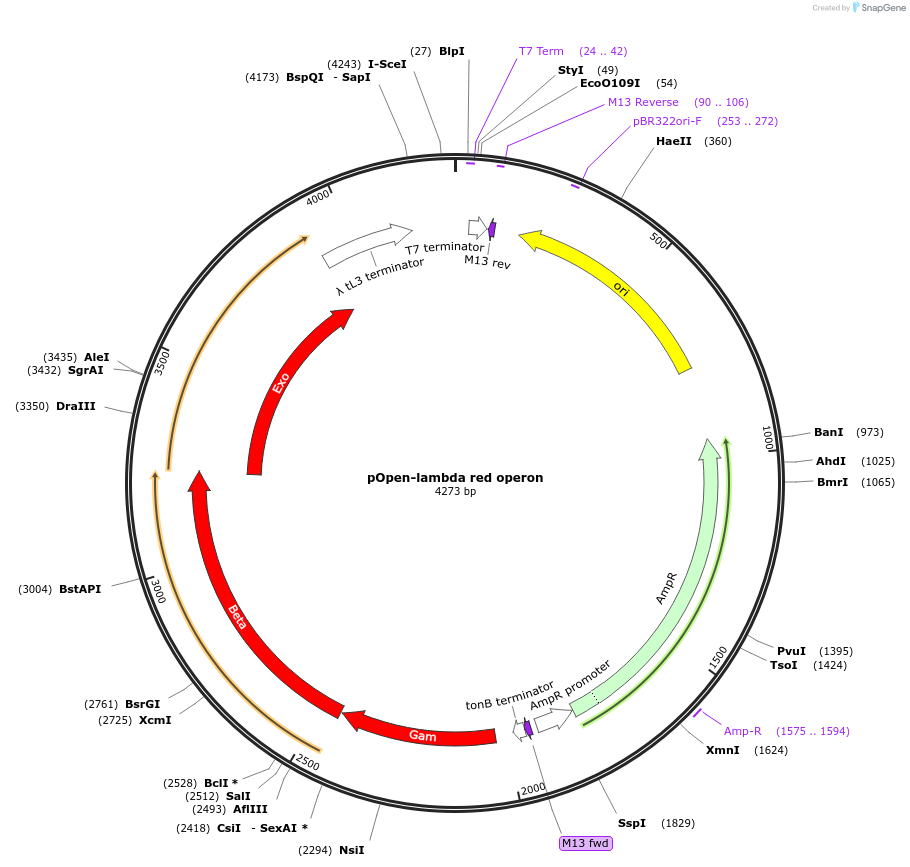
pOpen-lambda red operon
Plasmid#165521PurposeOperon containing Exo, Bet, and Gam. To use the lambda red recombineering system to modify your target DNA.DepositorInsertlambda red operon
UseSynthetic BiologyAvailable SinceAug. 3, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
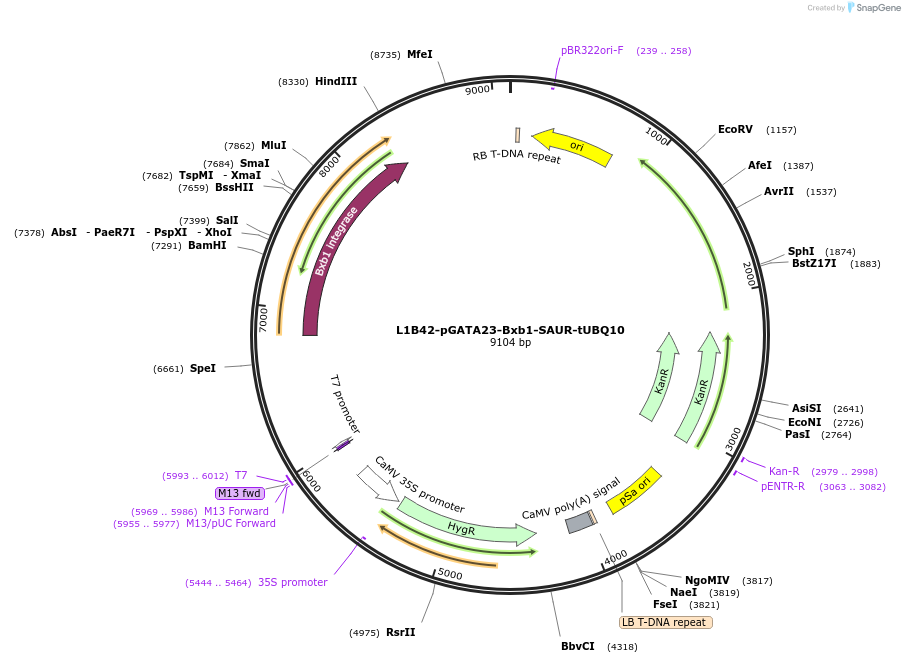
L1B42-pGATA23-Bxb1-SAUR-tUBQ10
Plasmid#219705PurposeLevel 1 pGATA23-Bxb1-SAUR-tUBQ10, for lateral root specific expression of Bxb1 integrase with SAUR tagDepositorInsertpGATA23:Bxb1:SAUR:tUBQ10
ExpressionPlantAvailable SinceOct. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
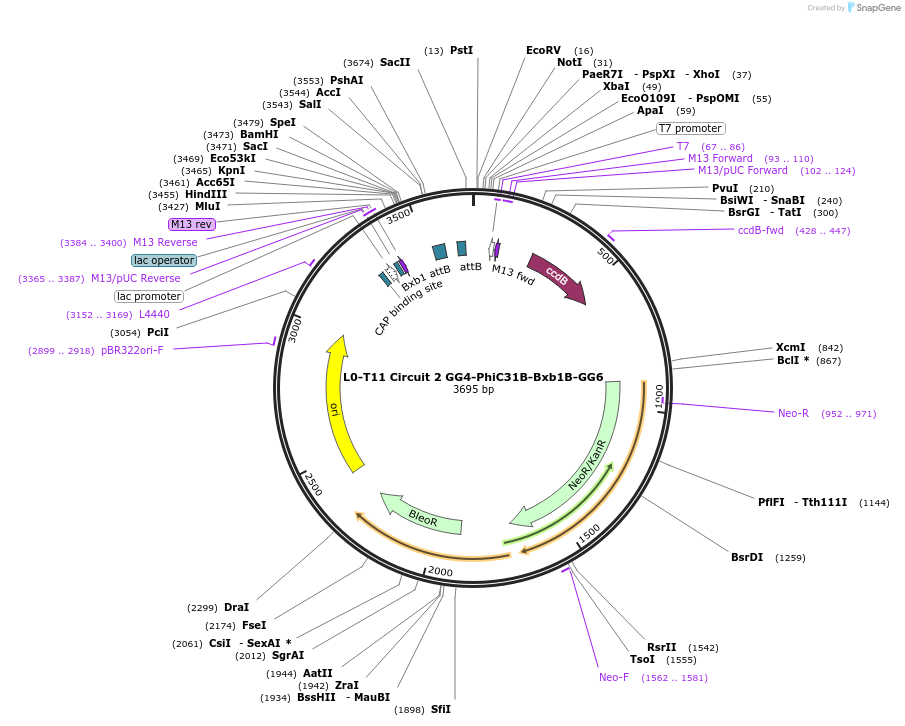
L0-T11 Circuit 2 GG4-PhiC31B-Bxb1B-GG6
Plasmid#219694PurposeLevel 0 to construct history-dependent target T2BPDepositorInsertGG4-PhiC31B-Bxb1B-GG6
UseSynthetic BiologyAvailable SinceSept. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
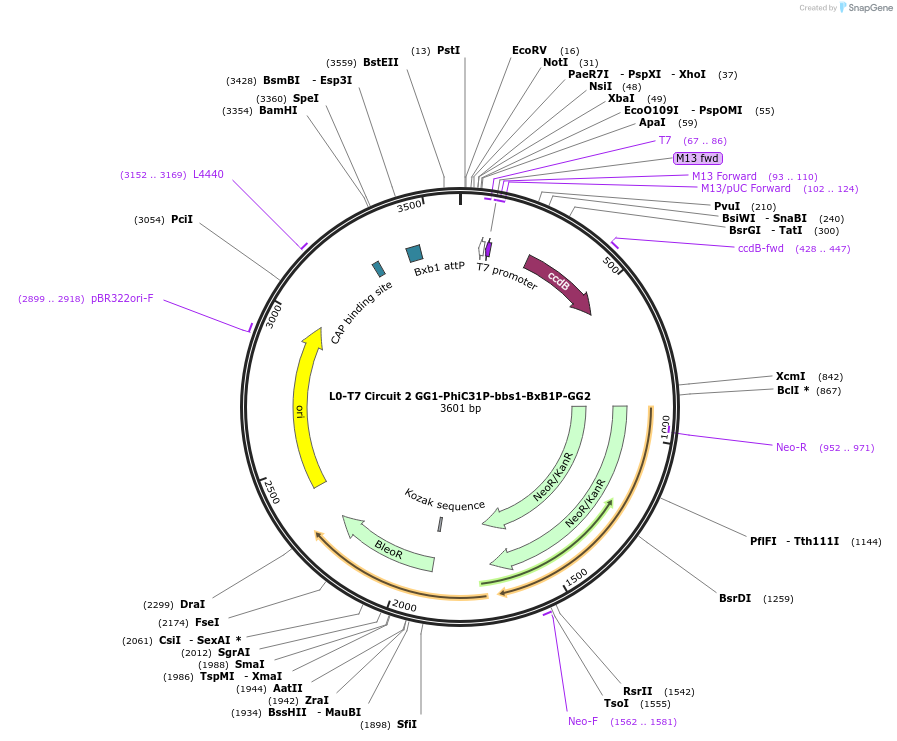
L0-T7 Circuit 2 GG1-PhiC31P-bbs1-BxB1P-GG2
Plasmid#219692PurposeLevel 0 to construct history-dependent target T2BPDepositorInsertGG1-PhiC31P-bbs1-BxB1P-GG2
UseSynthetic BiologyAvailable SinceSept. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
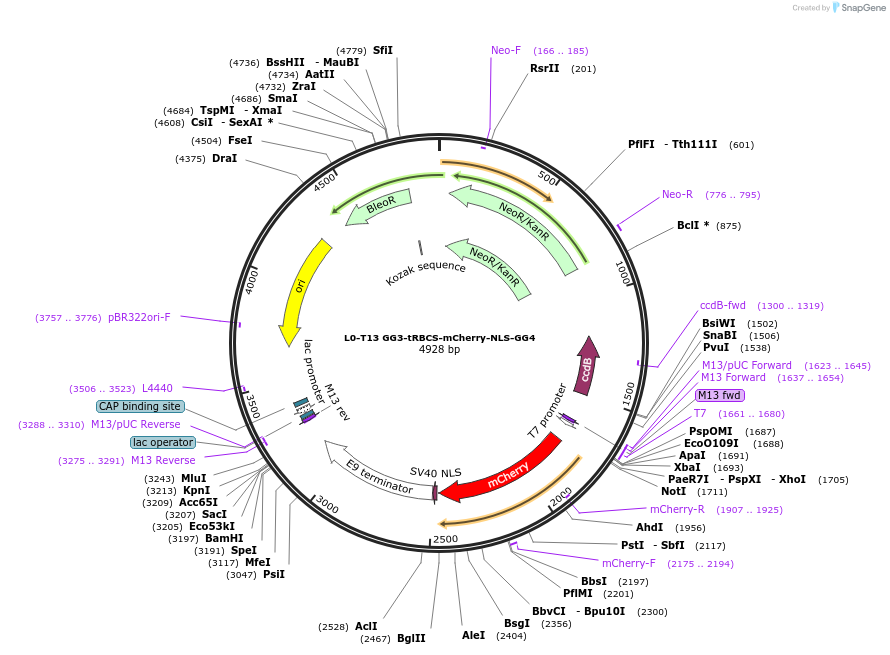
L0-T13 GG3-tRBCS-mCherry-NLS-GG4
Plasmid#219696PurposeLevel 0 to construct history-dependent targetDepositorInsertGG3-tRBCS-mCherry-NLS-GG4
UseSynthetic BiologyAvailable SinceJune 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
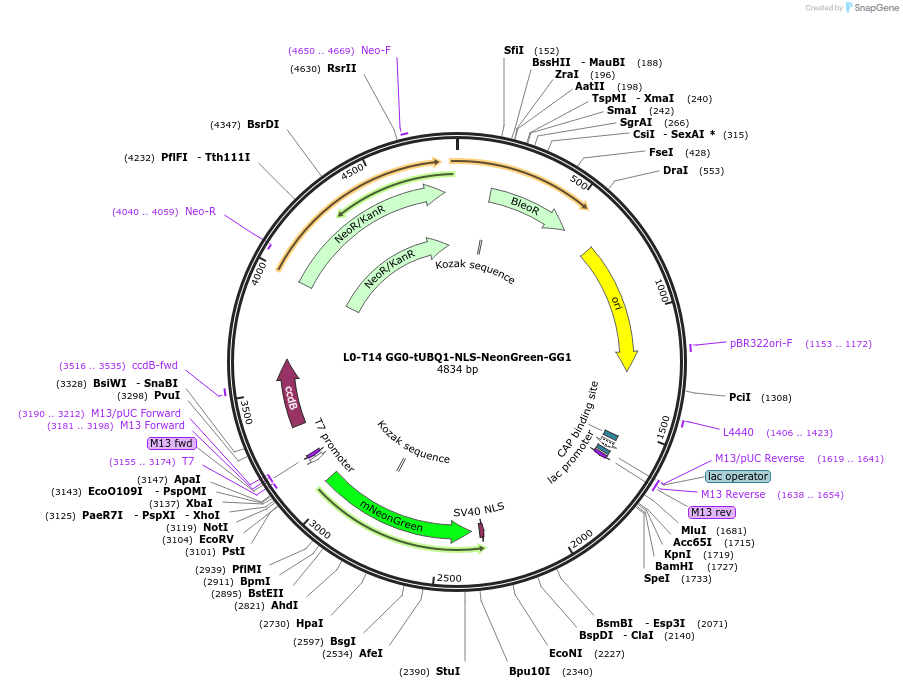
L0-T14 GG0-tUBQ1-NLS-NeonGreen-GG1
Plasmid#219697PurposeLevel 0 to construct history-dependent targetDepositorInsertGG0-tUBQ1-NLS-NeonGreen-GG1
UseSynthetic BiologyAvailable SinceJune 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
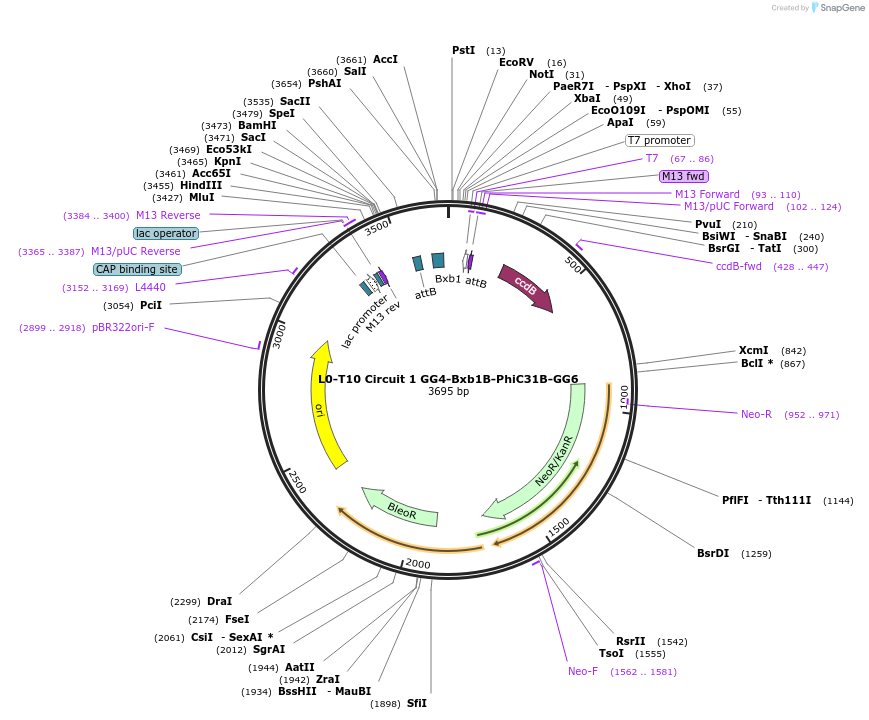
L0-T10 Circuit 1 GG4-Bxb1B-PhiC31B-GG6
Plasmid#219693PurposeLevel 0 to construct history-dependent target T2PBDepositorInsertGG4-Bxb1B-PhiC31B-GG6
UseSynthetic BiologyAvailable SinceJune 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
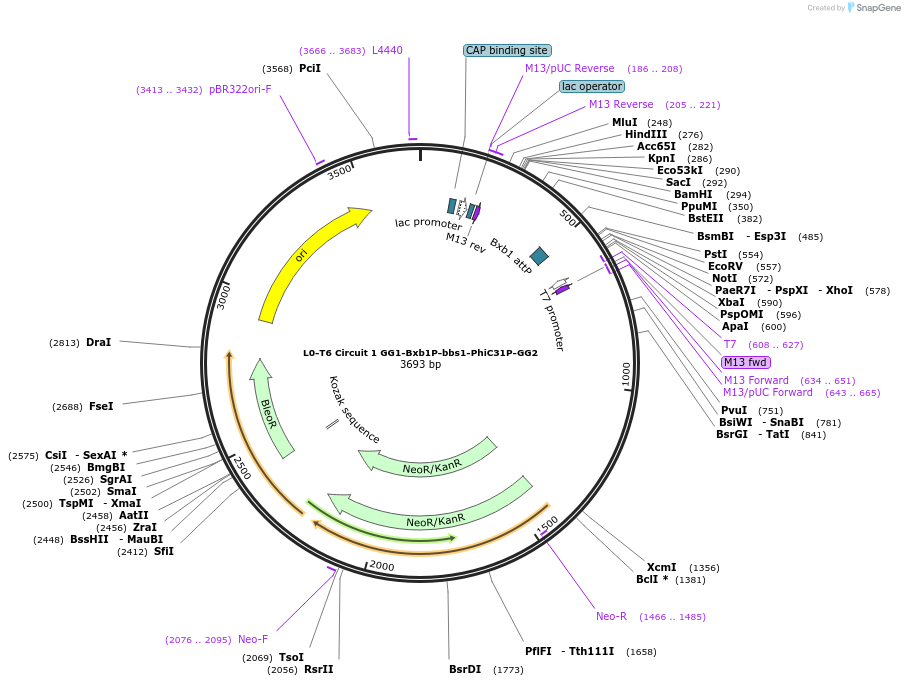
L0-T6 Circuit 1 GG1-Bxb1P-bbs1-PhiC31P-GG2
Plasmid#219691PurposeLevel 0 to construct history-dependent target T2PBDepositorInsertGG1-Bxb1P-bbs1-PhiC31P-GG2
UseSynthetic BiologyAvailable SinceJune 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
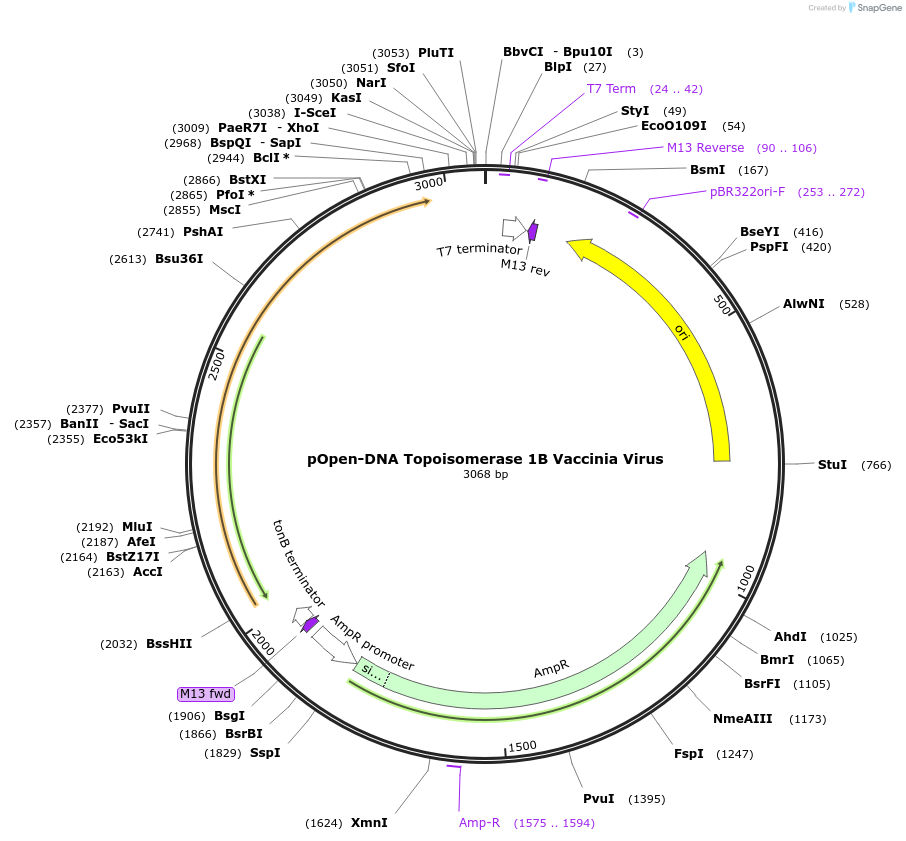
pOpen-DNA Topoisomerase 1B Vaccinia Virus
Plasmid#165510PurposeUsed in TOPO cloning. Recognizes the DNA sequence 5'-(C/T)CCTT-3' and digests double stranded DNA at this sequence.DepositorInsertDNA Topoisomerase 1B Vaccinia Virus
UseSynthetic BiologyAvailable SinceJuly 30, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
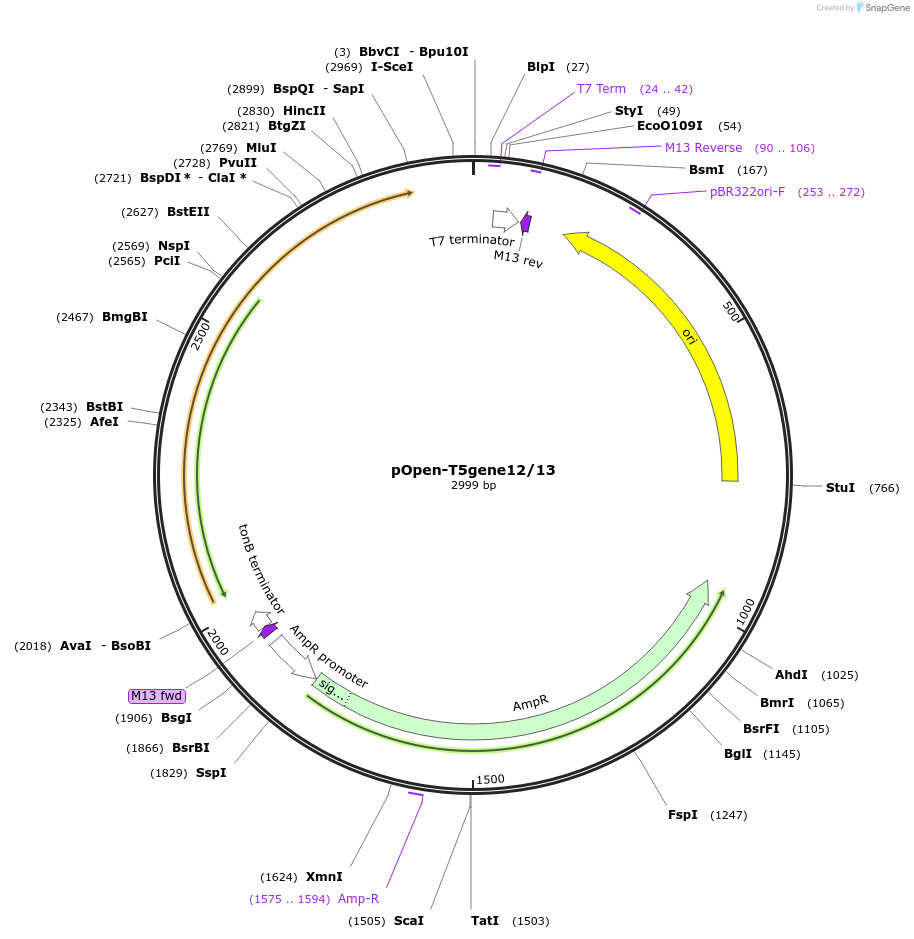
pOpen-T5gene12/13
Plasmid#165559PurposeDouble-stranded DNA specific exonuclease and single-stranded DNA endonuclease. Initiates at the 5' termini of linear or nicked double-stranded DNA. Cleaves linear or nicked double-stranded DNA in the 5' to 3' direction.DepositorInsertT5 Exonuclease
UseSynthetic BiologyAvailable SinceAug. 16, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
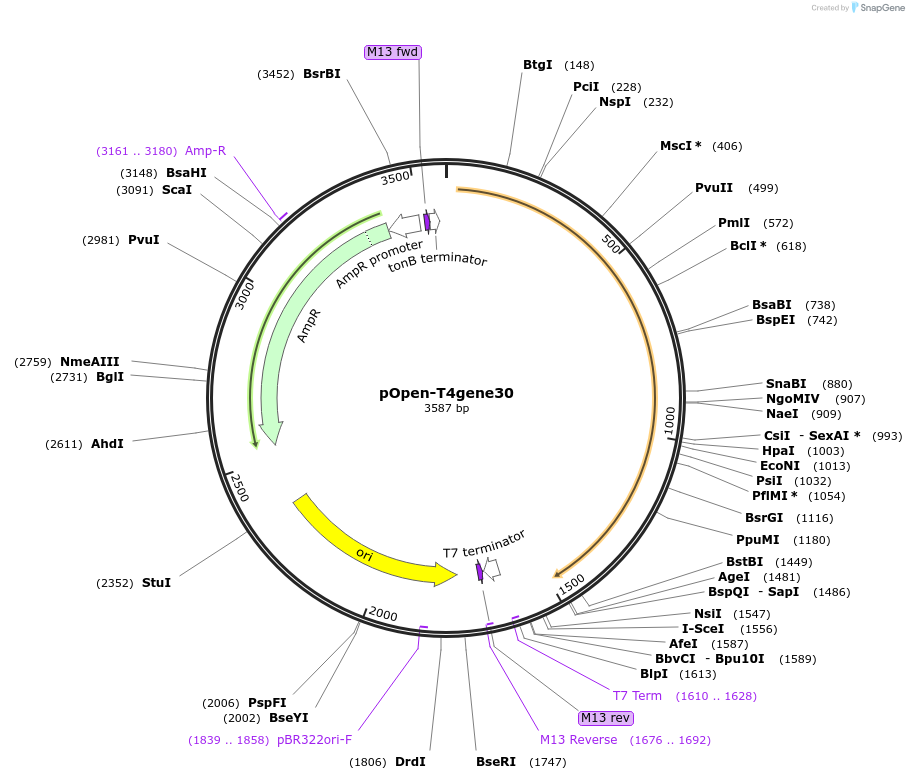
pOpen-T4gene30
Plasmid#165554PurposeLigase derived from T4 bacteriophage expressed in the early stage of lytic development. Has been implicated in T4 DNA synthesis and genetic recombination. May also play a role in T4 DNA repair.DepositorInsertT4-DNA Ligase
UseSynthetic BiologyAvailable SinceAug. 16, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
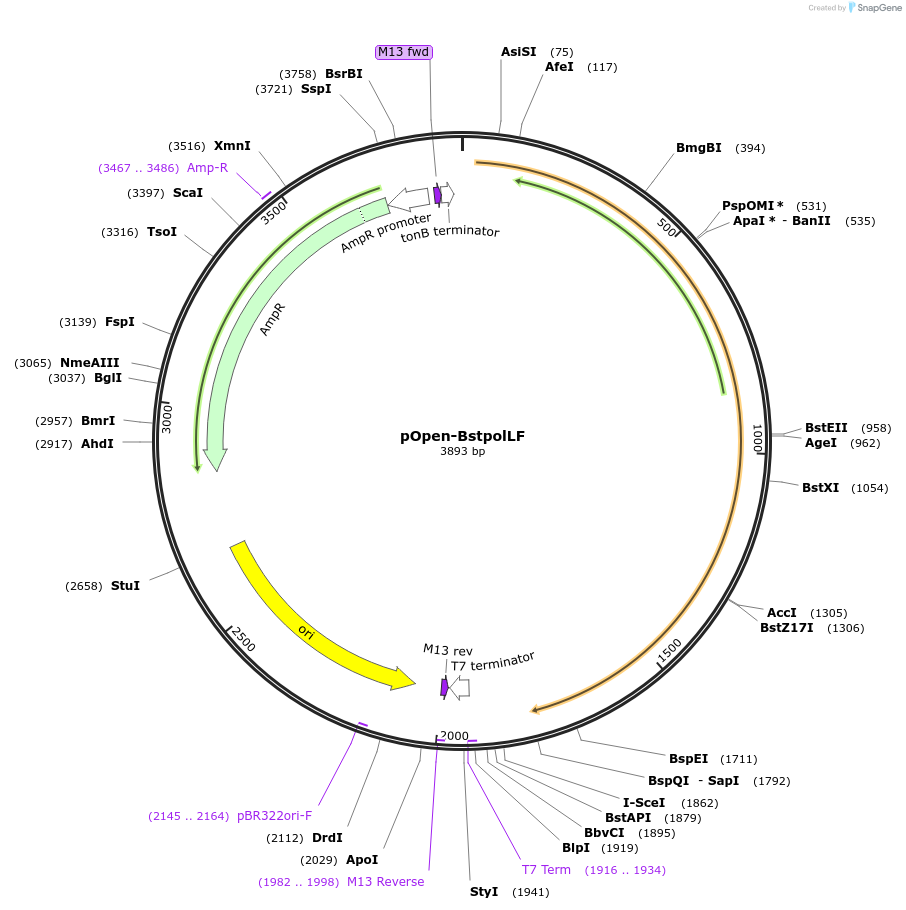
pOpen-BstpolLF
Plasmid#165553PurposeFragment retains 5'-3' polymerase activity from full length Bst DNA Polymerase, while lacking 5'-3' exonuclease activity. Suitable for applications requiring thermophilic strand displacement.DepositorInsertBst DNA Polymerase, Large Fragment
UseSynthetic BiologyAvailable SinceAug. 16, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
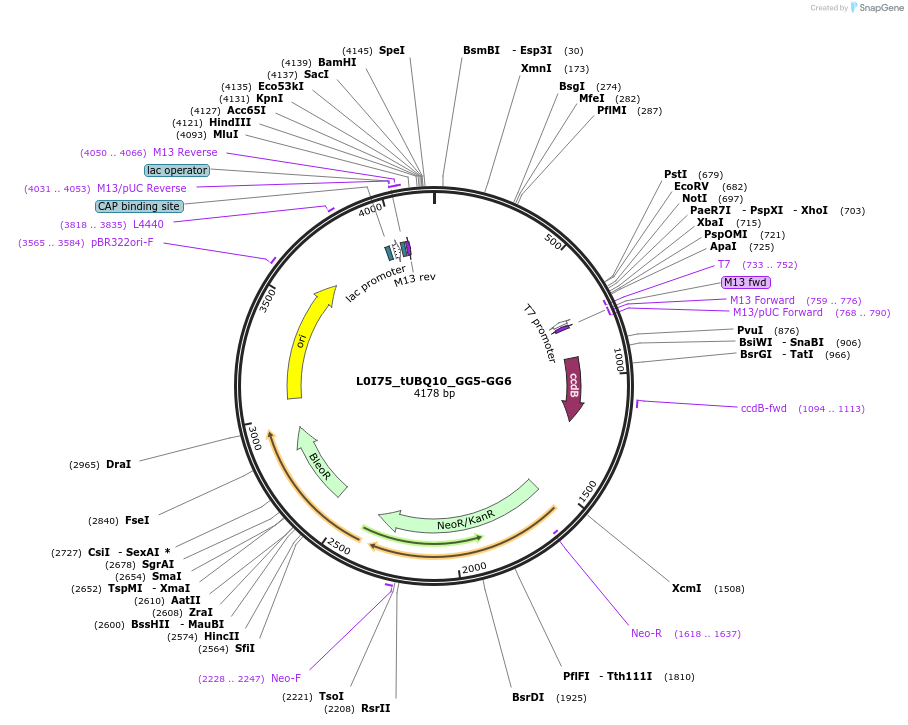
L0I75_tUBQ10_GG5-GG6
Plasmid#219714PurposeLevel 0 tUBQ10 terminator, GG5 and GG6 BsaI sites for golden gate assembly.DepositorInsertGG5-tUBQ10-GG6
UseSynthetic BiologyMutationWT terminator with added golden gate linkersAvailable SinceJuly 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
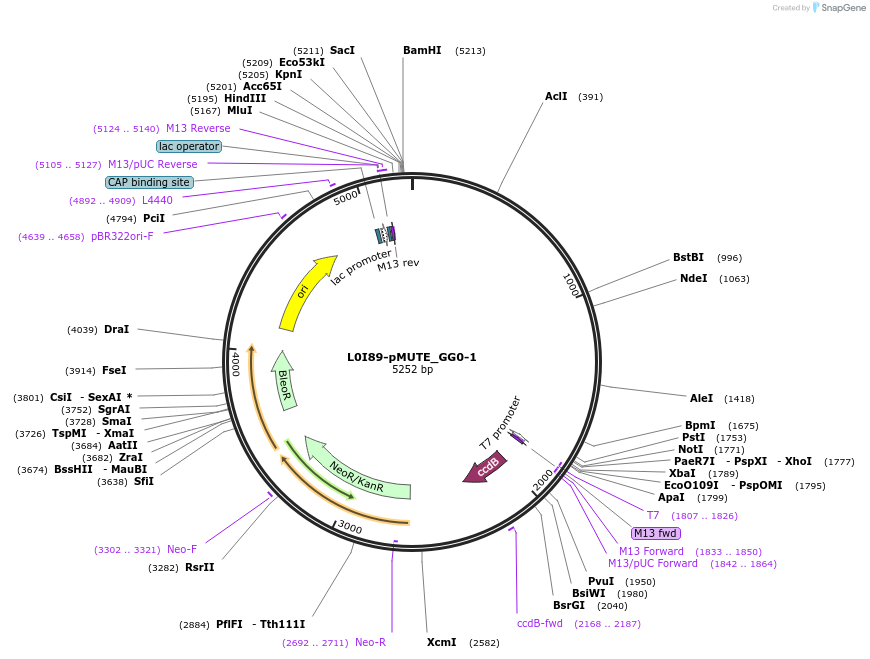
L0I89-pMUTE_GG0-1
Plasmid#219708PurposeLevel 0 pMUTE promoter, GG0 and GG1 BsaI sites for golden gate assembly.DepositorInsertGG0-pMUTE-GG1
UseSynthetic BiologyMutationWT promoter with added golden gate linkersAvailable SinceJune 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
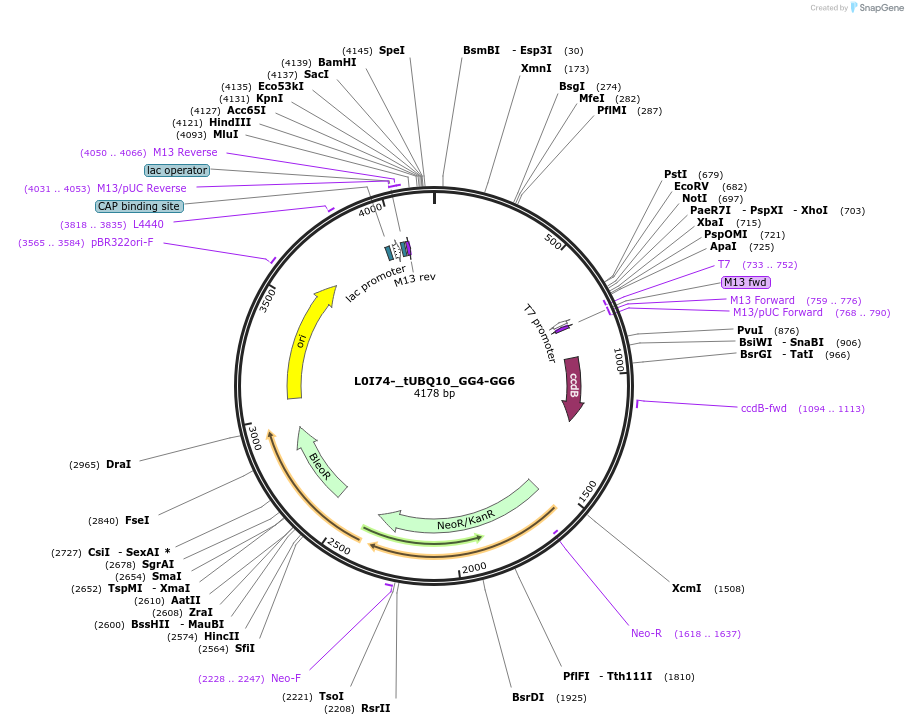
L0I74-_tUBQ10_GG4-GG6
Plasmid#219713PurposeLevel 0 tUBQ10 terminator, GG4 and GG6 BsaI sites for golden gate assembly.DepositorInsertGG4-tUBQ10-GG6
UseSynthetic BiologyMutationWT terminator with added golden gate linkersAvailable SinceJune 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
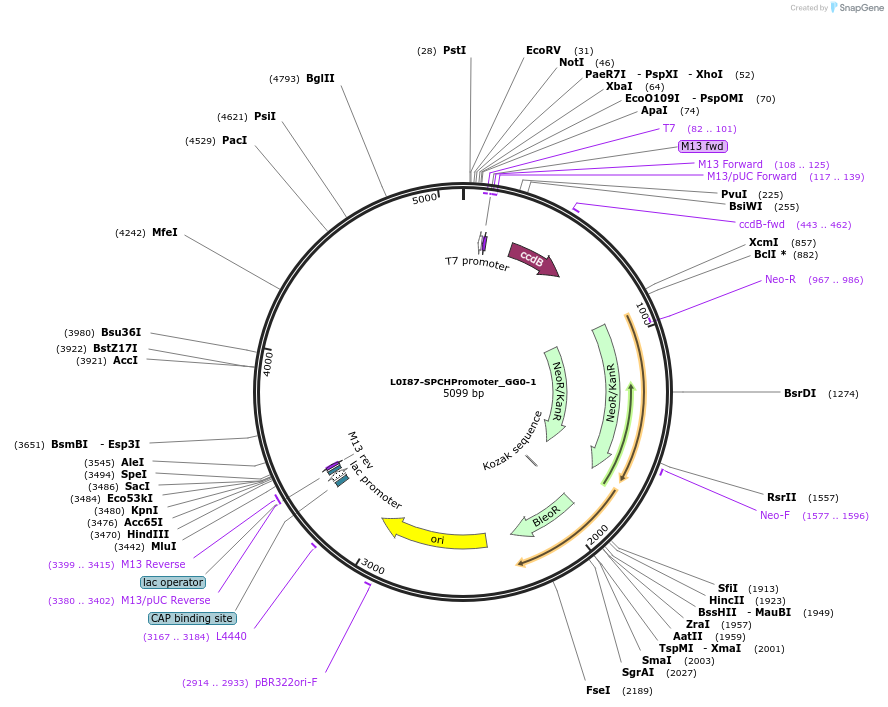
L0I87-SPCHPromoter_GG0-1
Plasmid#219707PurposeLevel 0 pSPCH promoter, GG0 and GG1 BsaI sites for golden gate assembly.DepositorInsertGG0-pSPCH-GG1
UseSynthetic BiologyMutationWT promoter with added golden gate linkersAvailable SinceJune 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
L0I80_Bxb1mut(noBsaI)_GG1-GG4
Plasmid#219715PurposeLevel 0 Bxb1 integrase with mutated BsaI sites, GG1 and GG4 BsaI sites for golden gate assembly.DepositorInsertGG1-Bxb1(noBsaI0)-GG4
UseSynthetic BiologyAvailable SinceJune 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
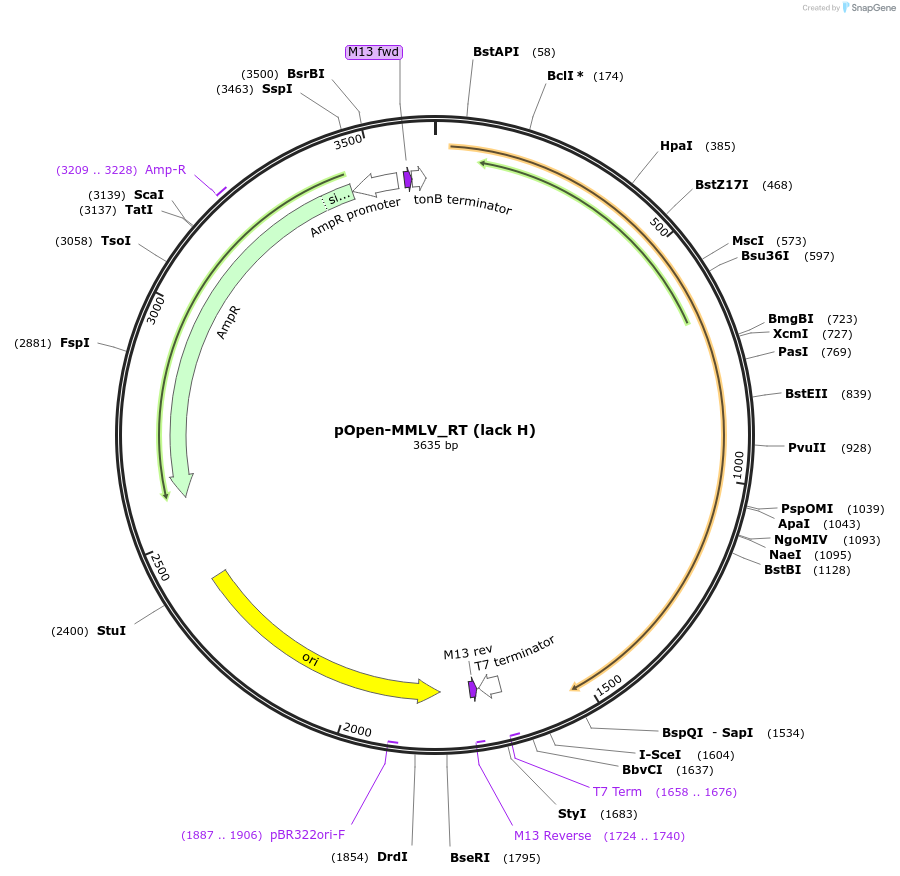
pOpen-MMLV_RT (lack H)
Plasmid#165556PurposeSingle 75 kDa monomer, cDNA synthesis; high enzyme activity and processivity. Comparable to SuperScriptII from Thermo Fisher.DepositorInsertMoloney Murine Leukemia Virus (MMLV) Reverse Transcriptase RNaseH - (lacking RNaseH domain)
UseSynthetic BiologyAvailable SinceAug. 16, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
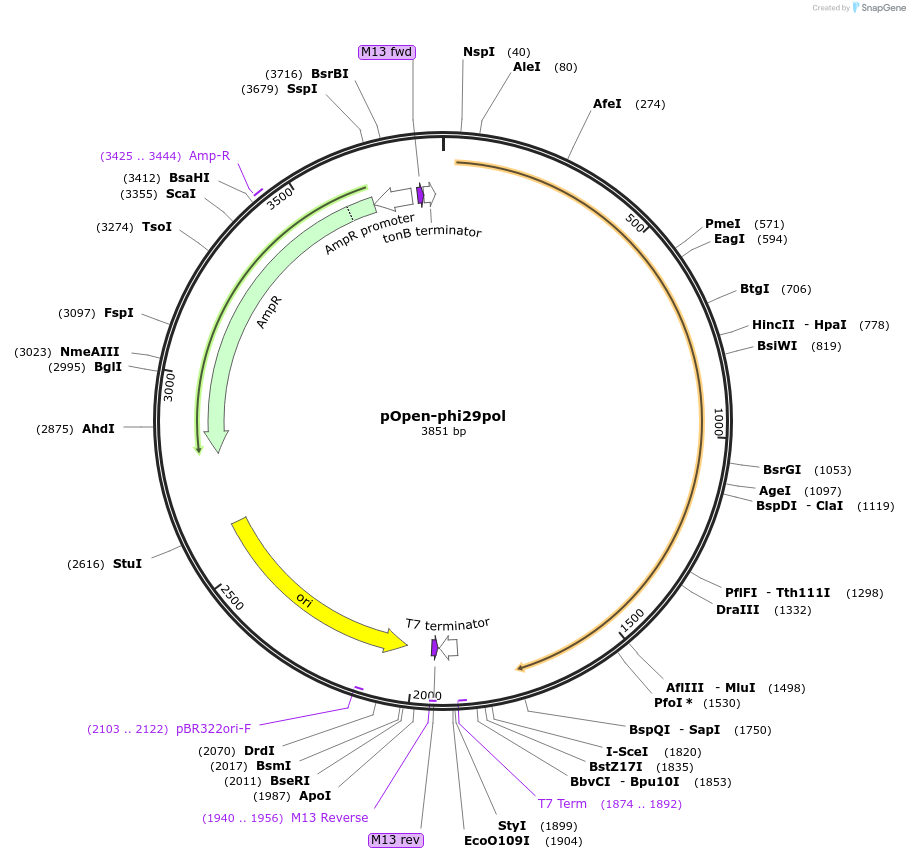
pOpen-phi29pol
Plasmid#165573PurposeDNA polymerase responsible for protein-primed viral DNA replication by strand displacement with high processivity and fidelity. Possesses three enzymatic activities: DNA synthesis (polymerase), primer terminal protein (TP) deoxynucleotidylation, and 3' to 5' exonuclease activity.DepositorInsertphi29 DNA Polymerase
UseSynthetic BiologyAvailable SinceAug. 24, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
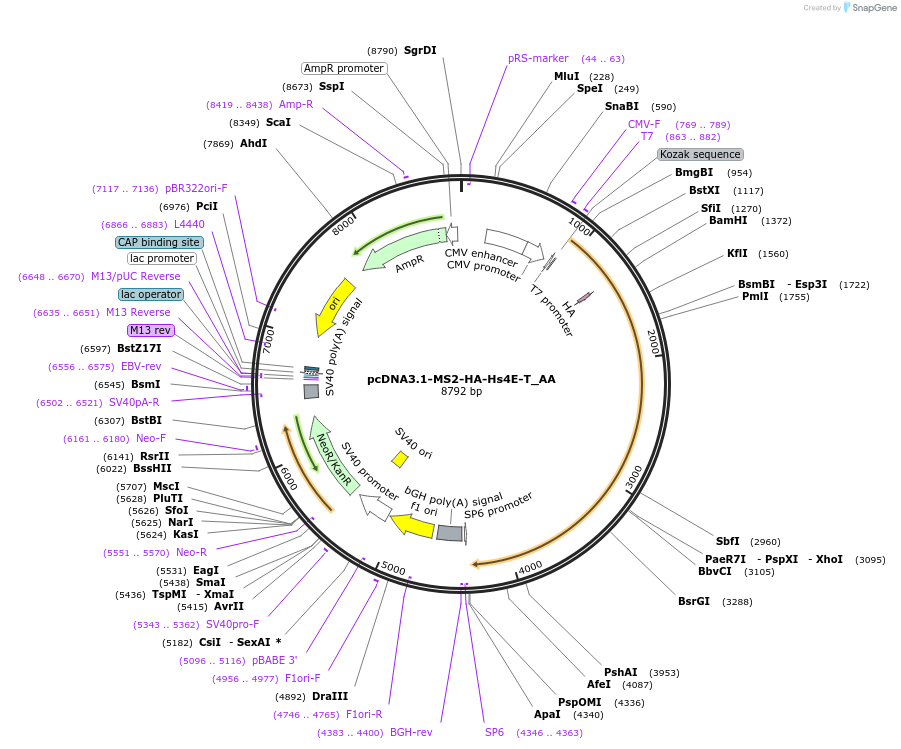
pcDNA3.1-MS2-HA-Hs4E-T_AA
Plasmid#148233PurposeMammalian Expression of Hs4E-TDepositorInsertHs4E-T (EIF4ENIF1 Human)
ExpressionMammalianAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
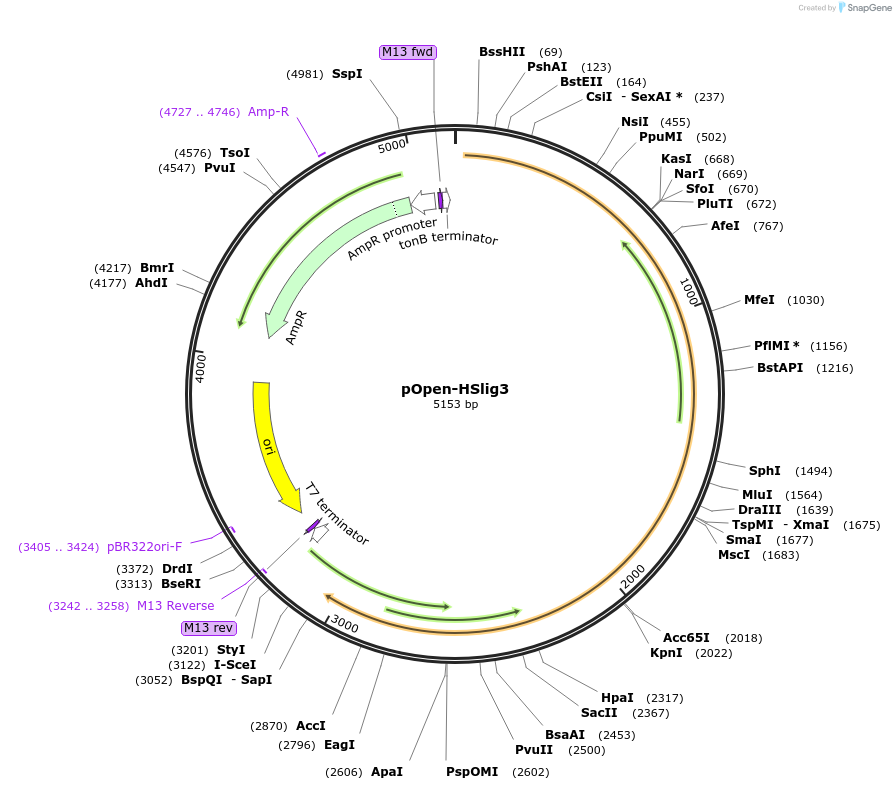
pOpen-HSlig3
Plasmid#165574PurposeRepairs single strand breaks in DNA efficiently. Unable to perform either blunt-end joining or AMP- dependent relaxation of supercoiled DNA (Elder, R.H. et al . , Bur. J. Biochem.. 203:53-58 (1992))DepositorInsertDNA Ligase III
UseSynthetic BiologyAvailable SinceAug. 24, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits