We narrowed to 20,812 results for: OMP
-
Plasmid#223424PurposeT-DNA vector for dLbCas12a-D156R mediated A to G base editing for monocot plants; TTTV PAM; LbCas12a-D156R-ABE and the crRNA were driven by separate ZmUbi1 promoter; BASTA for plants selection.DepositorInsertZmUbi-ecTadA8e-dLbCas12a-D156R-ZmUbi-crRNA scaffold
UseCRISPRTagsNLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
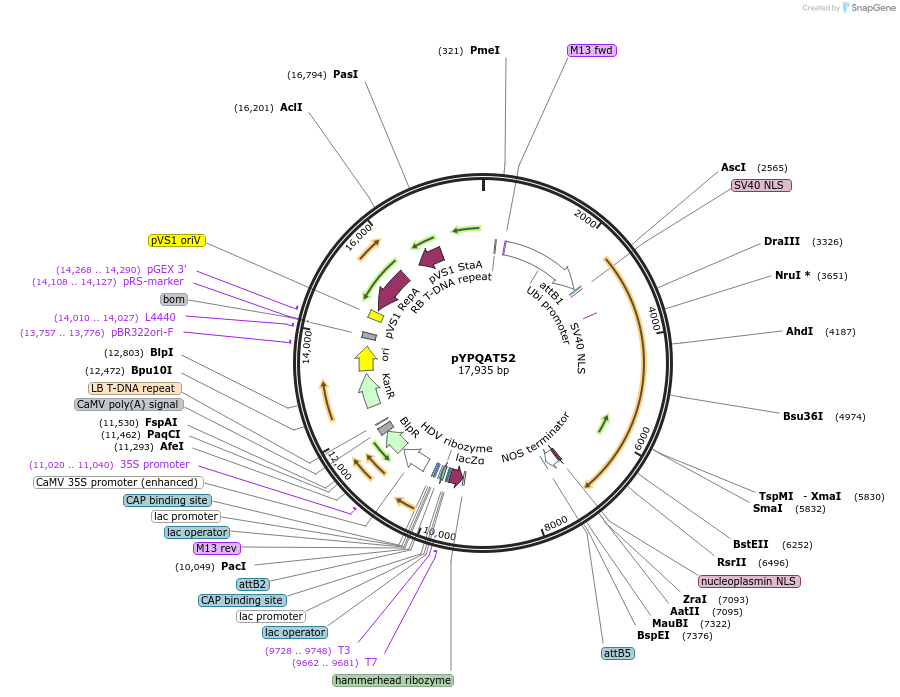
pYPQAT53
Plasmid#223425PurposeT-DNA vector for dLbCas12a-D156R mediated A to G base editing for monocot plants; TTTV PAM; LbCas12a-D156R-ABE and the crRNA were driven by separate ZmUbi1 promoter; Hygromycin for plants selection.DepositorInsertZmUbi-ecTadA8e-dLbCas12a-D156R-ZmUbi-crRNA scaffold
UseCRISPRTagsNLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
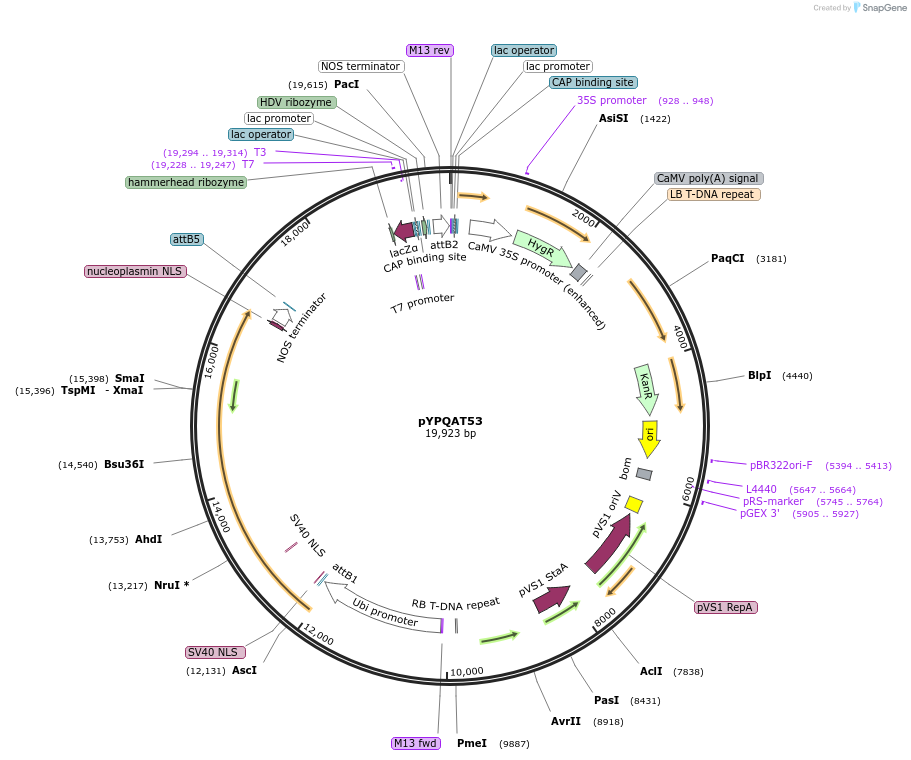
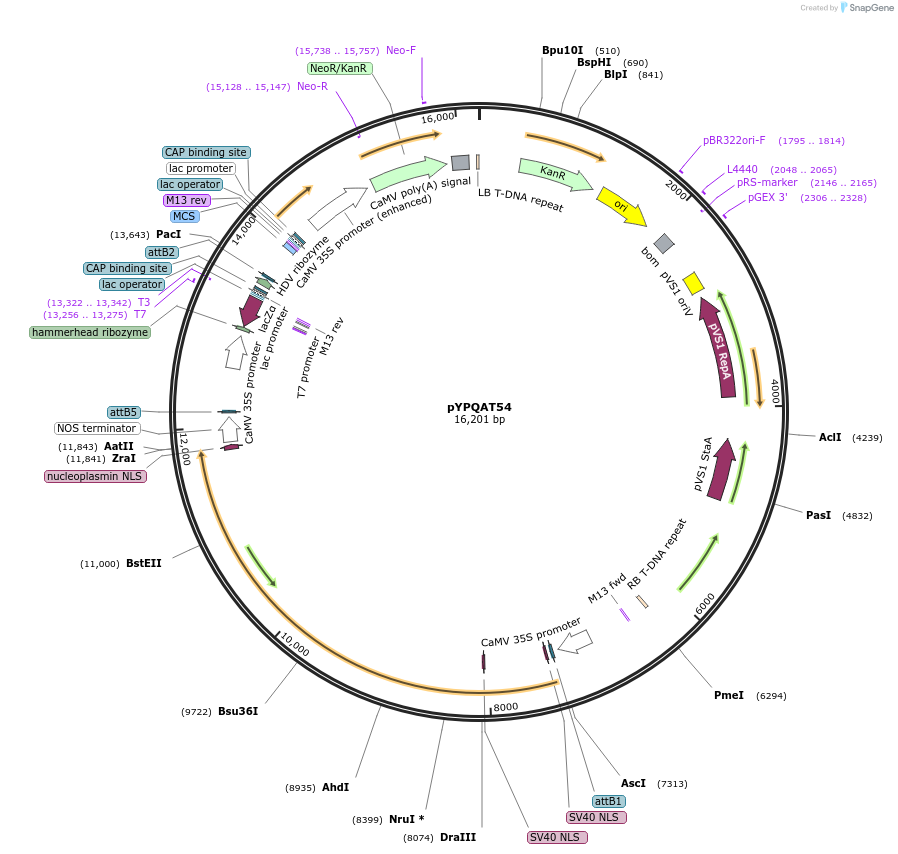
pYPQAT54
Plasmid#223426PurposeT-DNA vector for dLbCas12a-D156R mediated A to G base editing for monocot plants; TTTV PAM; LbCas12a-D156R-ABE and the crRNA were driven by separate 2x35s promoter; Kanamycin for plants selection.DepositorInsert2x35s-ecTadA8e-dLbCas12a-D156R-2x35s-crRNA scaffold
UseCRISPRTagsNLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
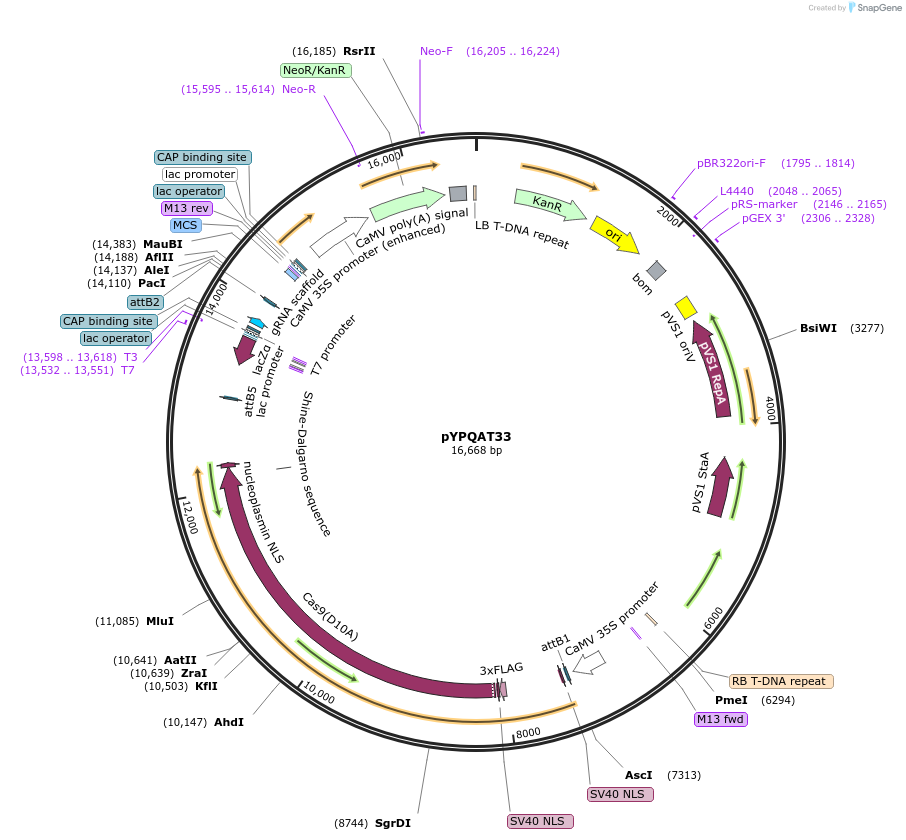
pYPQAT33
Plasmid#223405PurposeT-DNA vector for SpCas9-D10A based A-to-G base editing for dicot plants; NGG PAM; SpCas9D10A-ABE was driven by 2x35s and the sgRNA was driven by AtU3 promoter; Kanamycin for plants selection.DepositorInsert2x35s-ecTadA8e-SpCas9-D10A-AtU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
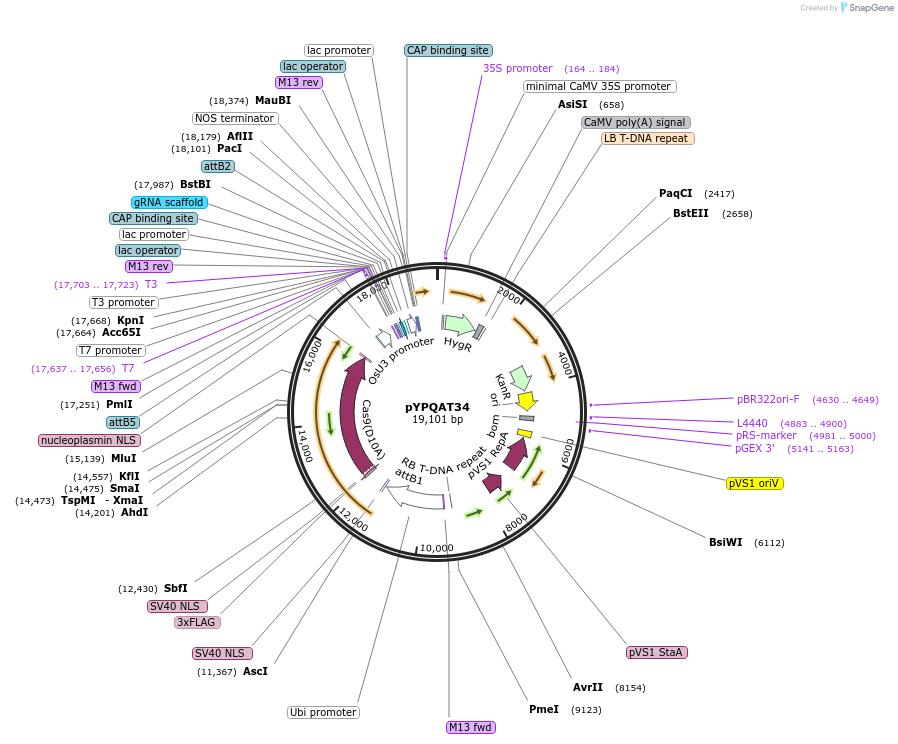
pYPQAT34
Plasmid#223406PurposeT-DNA vector for SpCas9-D10A based A-to-G base editing for monocot plants; NGG PAM; SpCas9D10A-ABE was driven by ZmUbi1 and the sgRNA was driven by OsU3 promoter; Hygromycin for plants selection.DepositorInsertZmUbi-ecTadA8e-SpCas9-D10A-OsU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
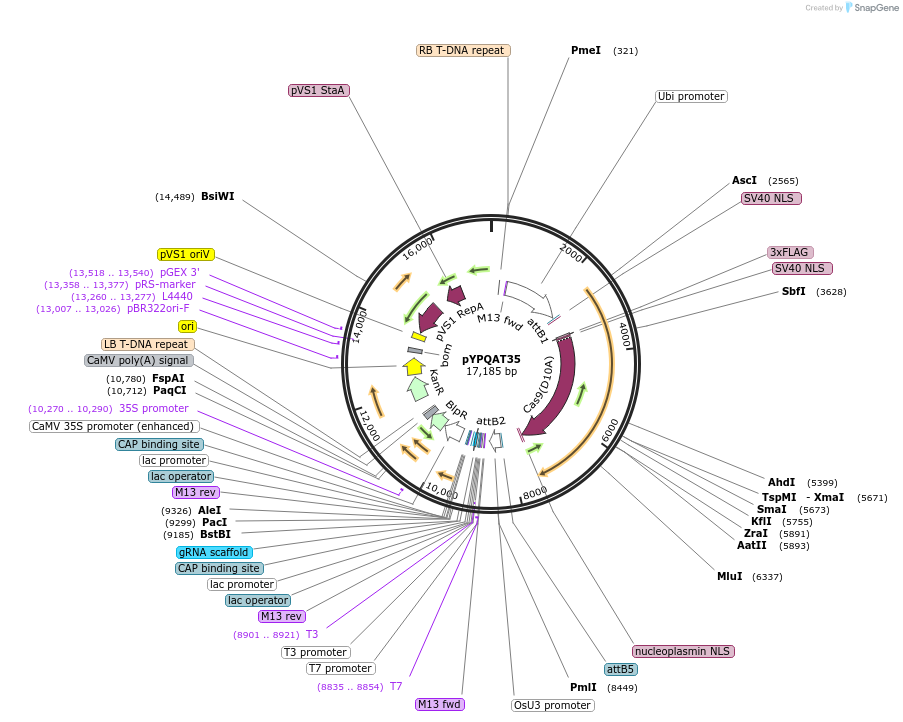
pYPQAT35
Plasmid#223407PurposeT-DNA vector for SpCas9-D10A based A-to-G base editing for monocot plants; NGG PAM; SpCas9D10A-ABE was driven by ZmUbi1 and the sgRNA was driven by OsU3 promoter; BASTA for plants selection.DepositorInsertZmUbi-ecTadA8e-SpCas9-D10A-OsU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
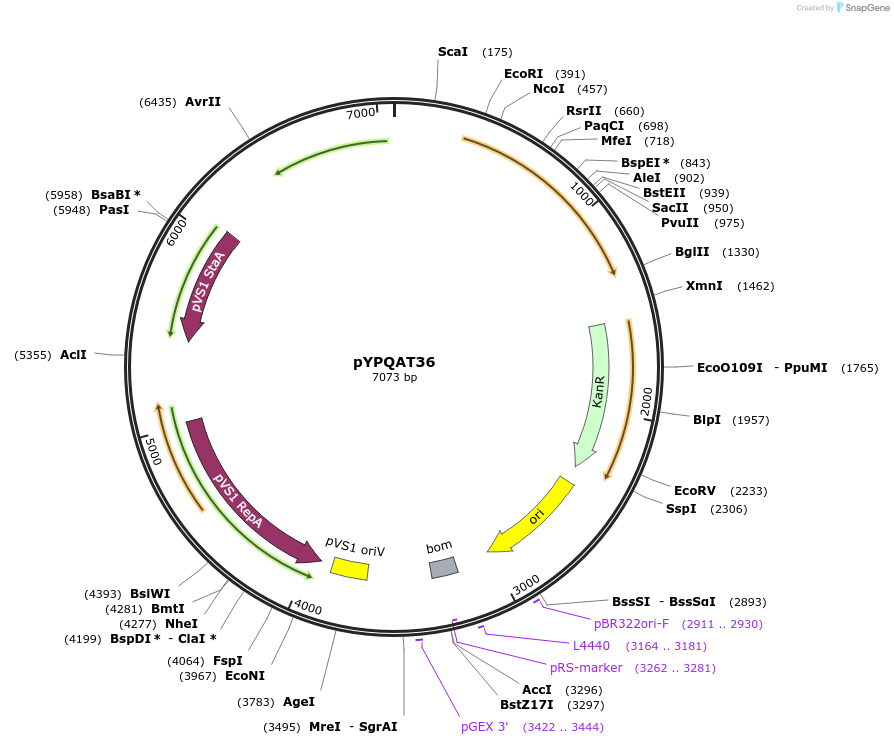
pYPQAT36
Plasmid#223408PurposeT-DNA vector for SpCas9-D10A based A-to-G base editing for monocot plants; NGG PAM; SpCas9D10A-ABE was driven by ZmUbi1 and the sgRNA was driven by ZmUbi1; Hygromycin for plants selection.DepositorInsertZmUbi-ecTadA8e-SpCas9-D10A-ZmUbi-gRNA scaffold-tRNA
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
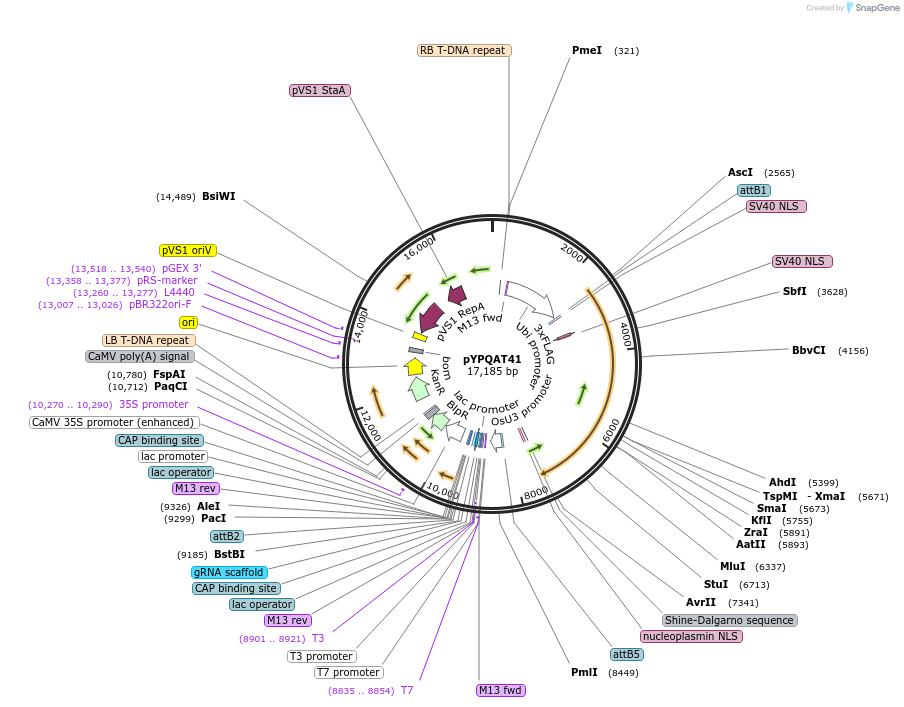
pYPQAT41
Plasmid#223413PurposeT-DNA vector for SpRY-D10A based A-to-G base editing for monocot plants; NA or NG PAM preference; SpRYD10A-ABE was driven by ZmUbi1 and the sgRNA was driven by OsU3 promoter; BASTA for plants select.DepositorInsertZmUbi-ecTadA8e-SpRY-D10A-OsU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
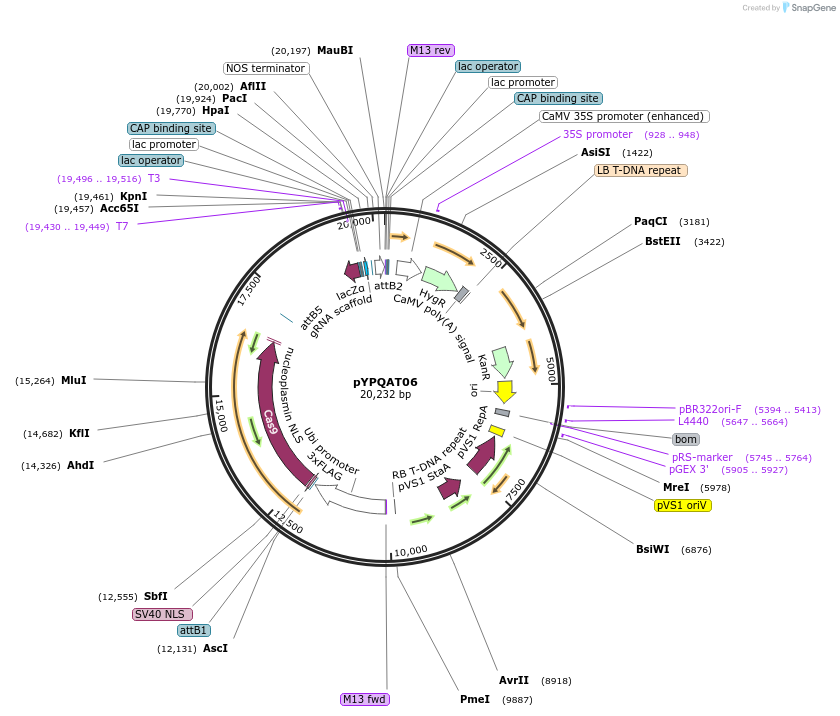
pYPQAT06
Plasmid#223378PurposeT-DNA vector for SpCas9 mediated mutagenesis for monocot plants; NGG PAM; Cas9 was driven by ZmUbi1 and the sgRNA was driven by ZmUbi1 promoter; Hygromycin for plants selection.DepositorInsertZmUbi-SpCas9-ZmUbi-gRNA scaffold-tRNA
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
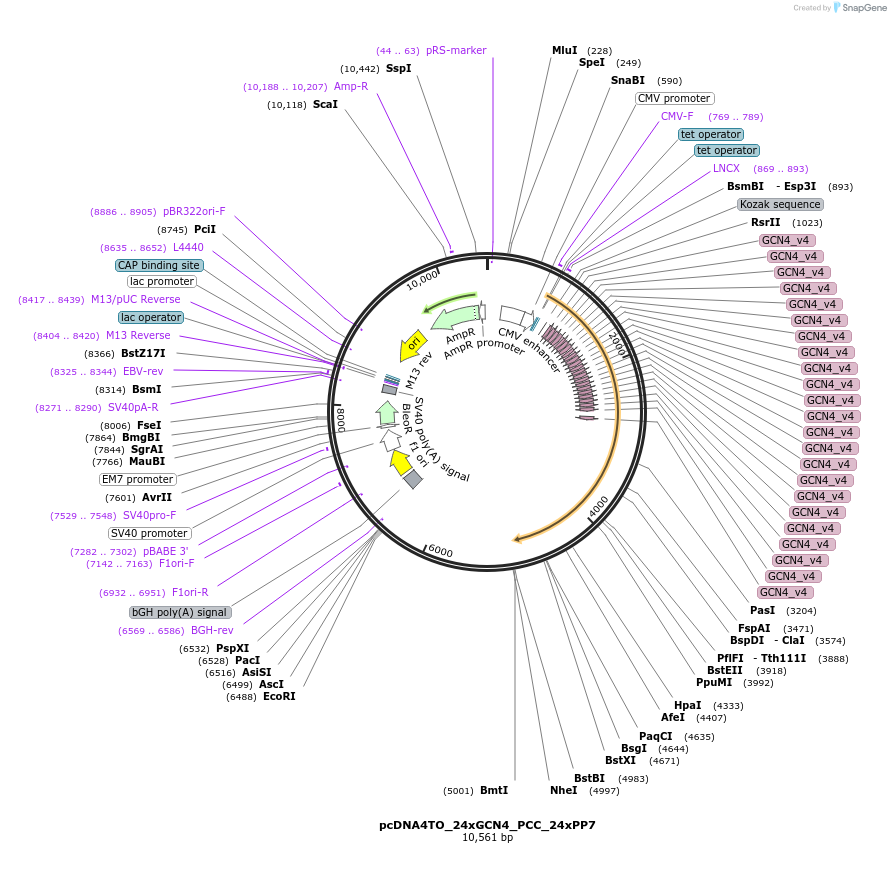
pcDNA4TO_24xGCN4_PCC_24xPP7
Plasmid#236094PurposeConstitutively encodes 24xGCN4-PCC-24xPP7 stem loops, for visualization of the Pcc mRNA.DepositorInsert24xGCN4-PCC-24xPP7
ExpressionMammalianAvailable SinceApril 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
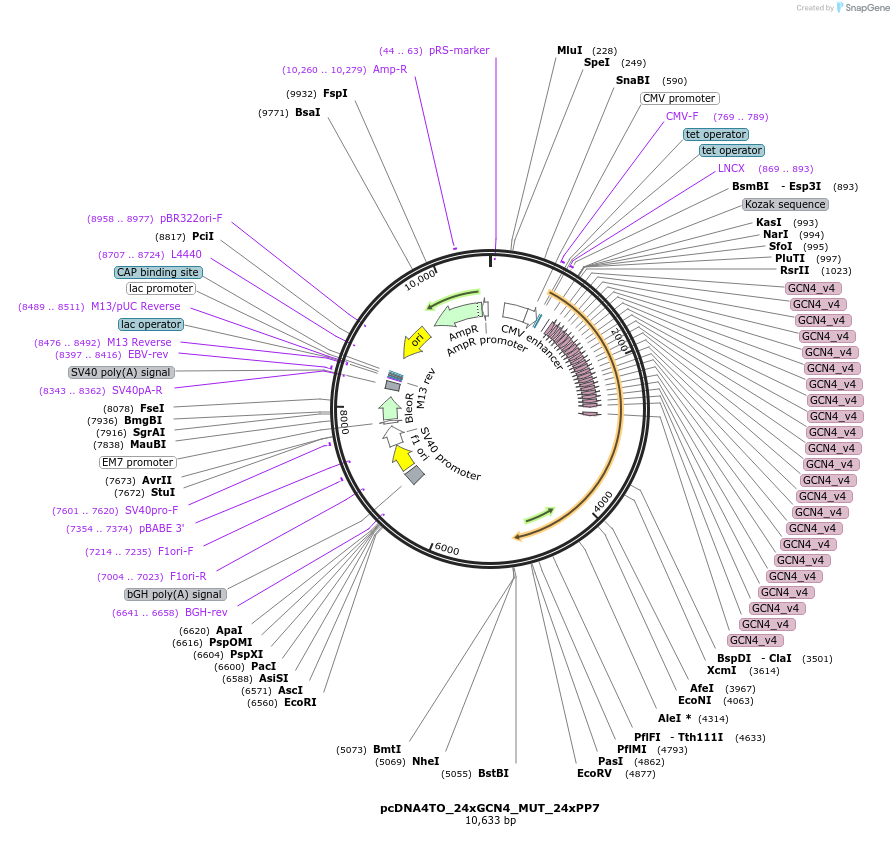
pcDNA4TO_24xGCN4_MUT_24xPP7
Plasmid#236093PurposeConstitutively encodes 24xGCN4-MUT-24xPP7 stem loops, for visualization of the Mut mRNA.DepositorInsert24xGCN4-MUT-24xPP7
ExpressionMammalianAvailable SinceApril 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
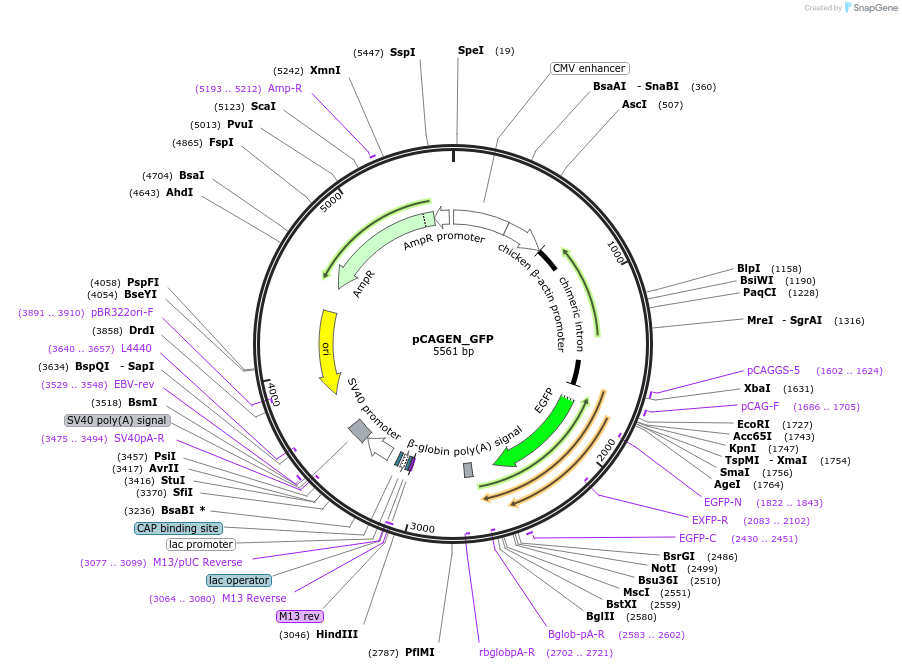
pCAGEN_GFP
Plasmid#235611PurposeEncodes GFP. For use in transfections as a negative control without epitope tags.DepositorInsertGFP
ExpressionMammalianAvailable SinceApril 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
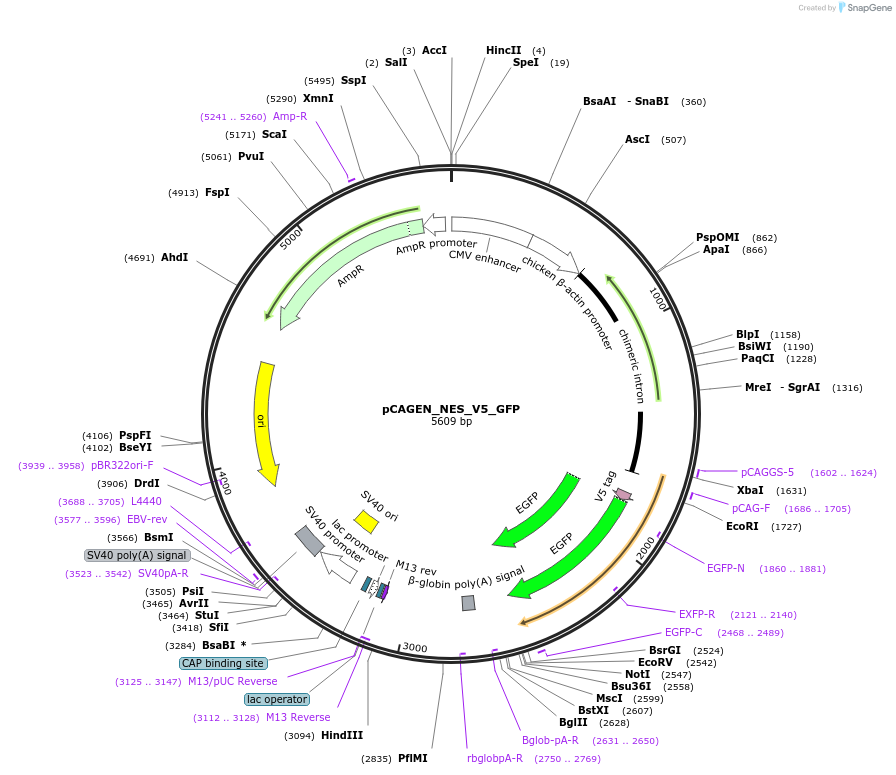
pCAGEN_NES_V5_GFP
Plasmid#235632PurposeEncodes V5-GFP with an N-terminal nuclear export sequence. Used as a no-ALIBi negative control.DepositorInsertGFP
TagsNES-V5ExpressionMammalianAvailable SinceApril 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
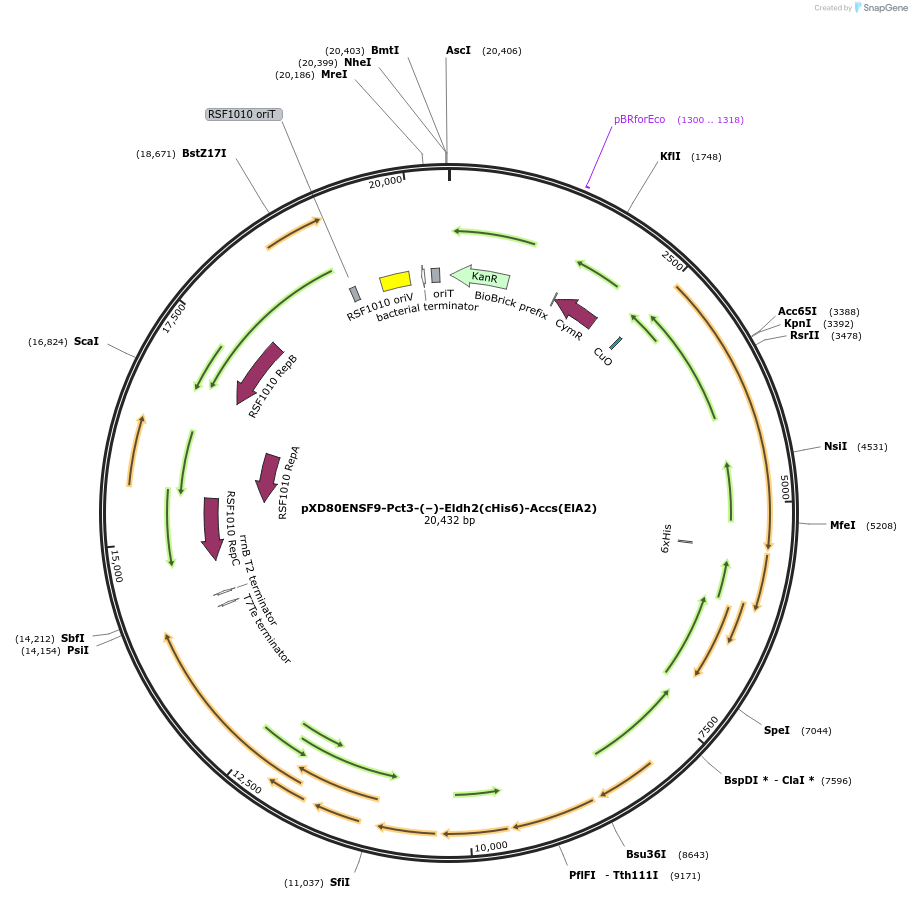
pXD80ENSF9-Pct3-(–)-Eldh2(cHis6)-Accs(ElA2)
Plasmid#233868PurposeExpresses His-tagged Eggerthella lenta A2 (–)-Eldh2 with a cumate inducible promoter in Gordonibacter urolithinfaciens DSM 27213 for protein purificationDepositorInsert(–)-Eldh2
TagsHisExpressionBacterialAvailable SinceApril 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
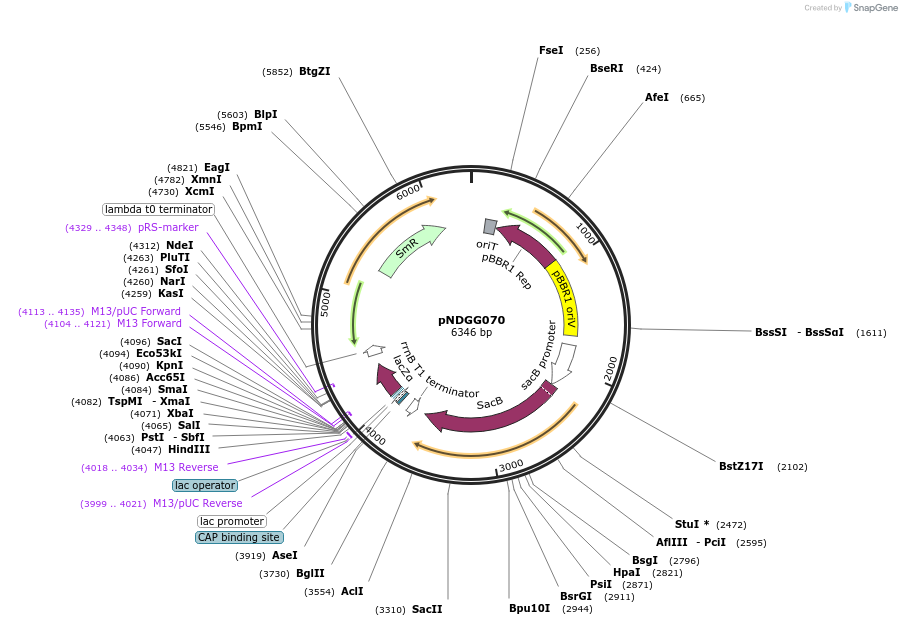
pNDGG070
Plasmid#231328PurposeBEVA Golden Gate cloning vector; BEVA2.0 Sucrose curable broad host range plasmid . L1-pBBR1-Sp-sacB-ELT4; GG assembly from pOGG004, pNDGG002, pOGG011, pNDGG008, and pOGG014, SpRDepositorTypeEmpty backboneExpressionBacterialAvailable SinceMarch 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
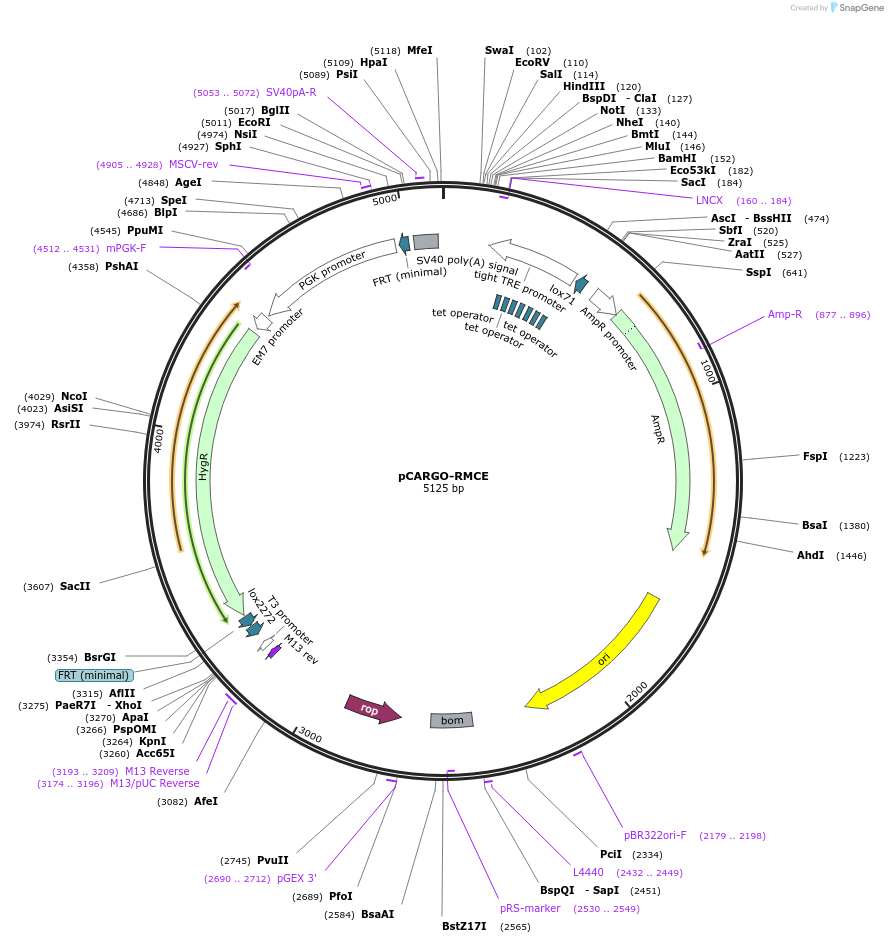
pCARGO-RMCE
Plasmid#233264PurposeBackbone plasmid used to clone lncRNA inserts in preparation for RMCEDepositorTypeEmpty backboneExpressionMammalianAvailable SinceMarch 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
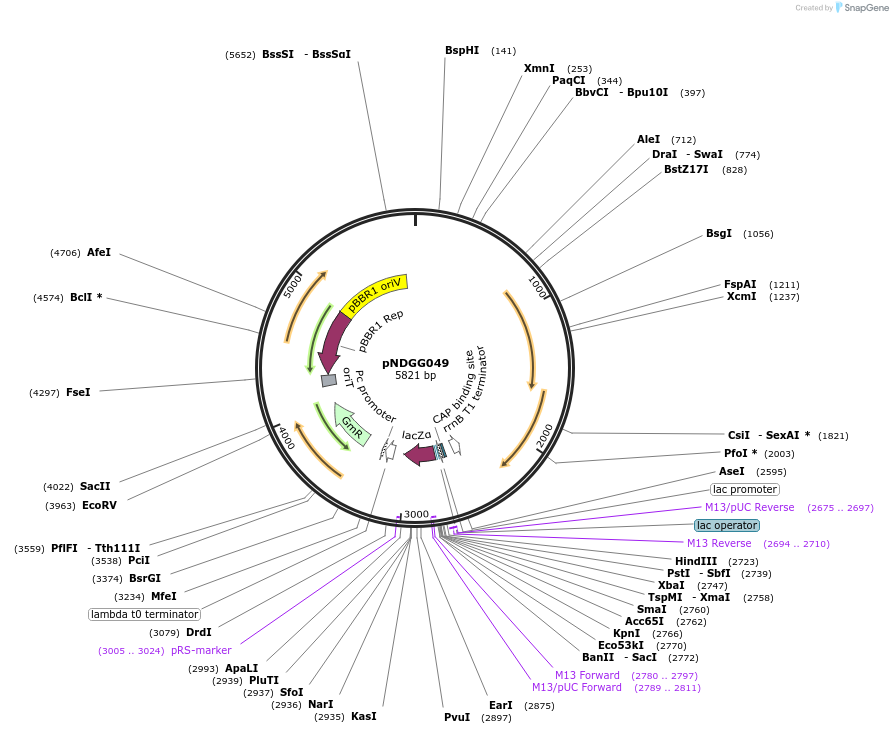
pNDGG049
Plasmid#231318PurposeBEVA Golden Gate cloning vector; BEVA2.0 Broad host range plasmid with stability. -pBBR1-Gm-par-ELT4; GG assembly from pOGG004, pOGG009, pOGG011, pOGG012, and pOGG014, GmRDepositorTypeEmpty backboneExpressionBacterialAvailable SinceMarch 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
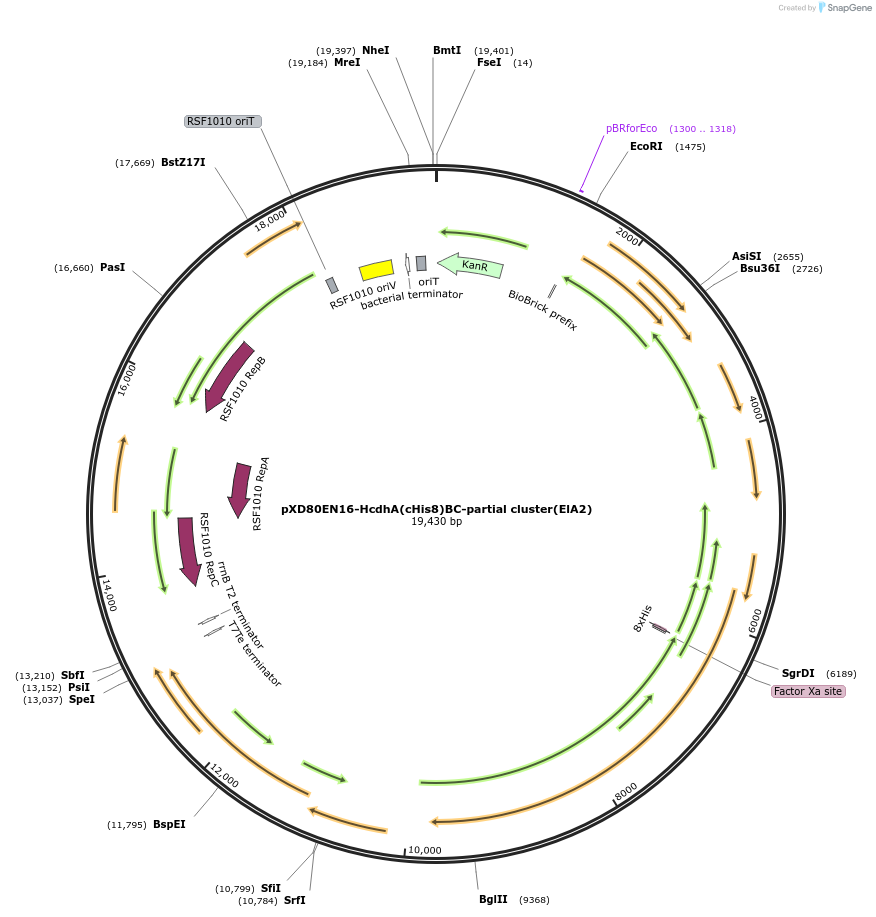
pXD80EN16-HcdhA(cHis8)BC-partial cluster(ElA2)
Plasmid#225328PurposeExpresses Eggerthella lenta A2 Hcdh partial gene cluster (inactive) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertHcdh partial gene cluster
TagsHis8ExpressionBacterialAvailable SinceMarch 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
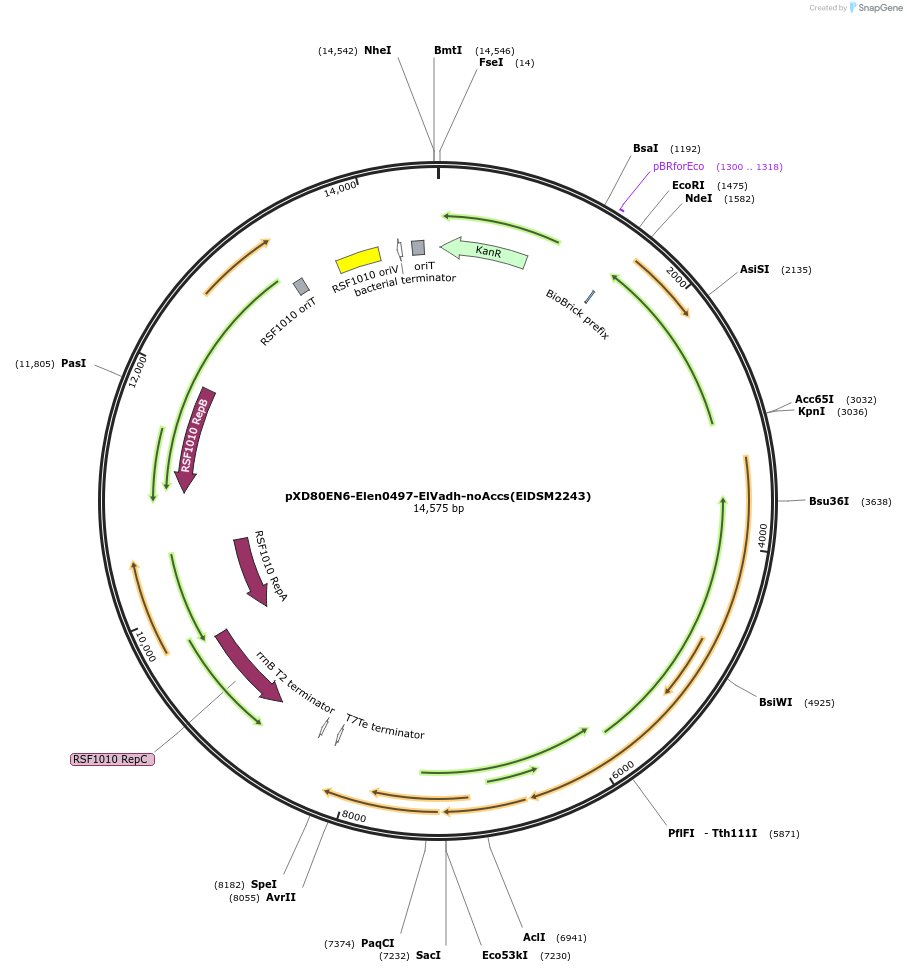
pXD80EN6-Elen0497-ElVadh-noAccs(ElDSM2243)
Plasmid#225314PurposeExpresses Eggerthella lenta DSM 2243 El Vadh without accessory genes (inactive) in Gordonibacter urolithinfaciens DSM 27213DepositorInsertEl Vadh (Elen_0497 Eggerthella lenta)
ExpressionBacterialAvailable SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
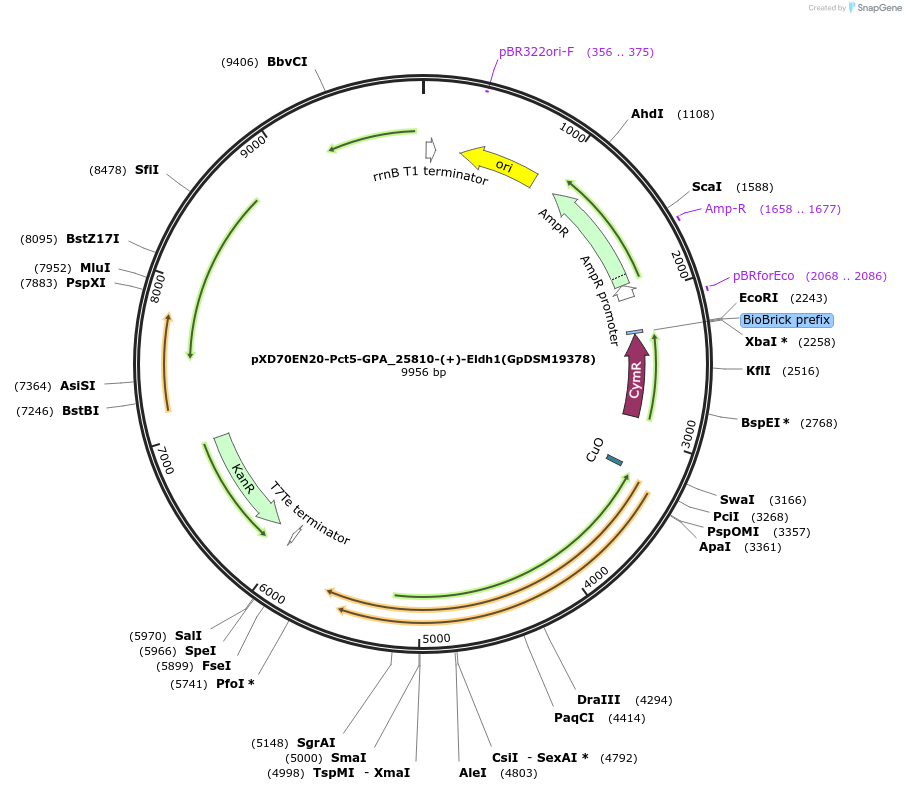
pXD70EN20-Pct5-GPA_25810-(+)-Eldh1(GpDSM19378)
Plasmid#225306PurposeExpresses Gordonibacter pamelaeae DSM 19378 (+)-Eldh1 in Eggerthella lenta DSM 2243 with a cumate inducible promoterDepositorInsert(+)-Eldh1
ExpressionBacterialAvailable SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
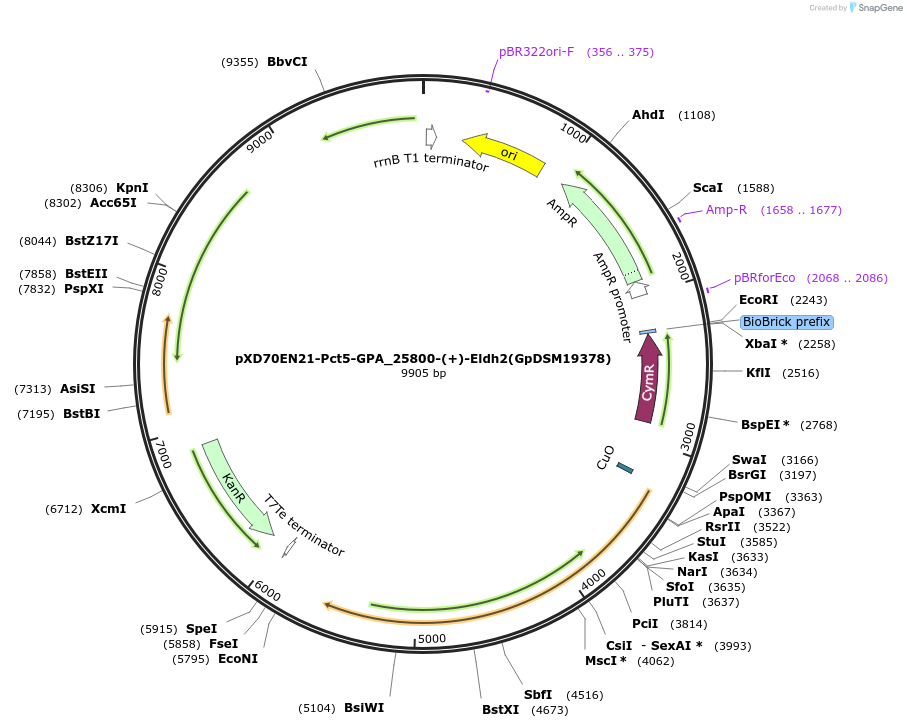
pXD70EN21-Pct5-GPA_25800-(+)-Eldh2(GpDSM19378)
Plasmid#225307PurposeExpresses Gordonibacter pamelaeae DSM 19378 (+)-Eldh2 in Eggerthella lenta DSM 2243 with a cumate inducible promoterDepositorInsert(+)-Eldh2
ExpressionBacterialAvailable SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
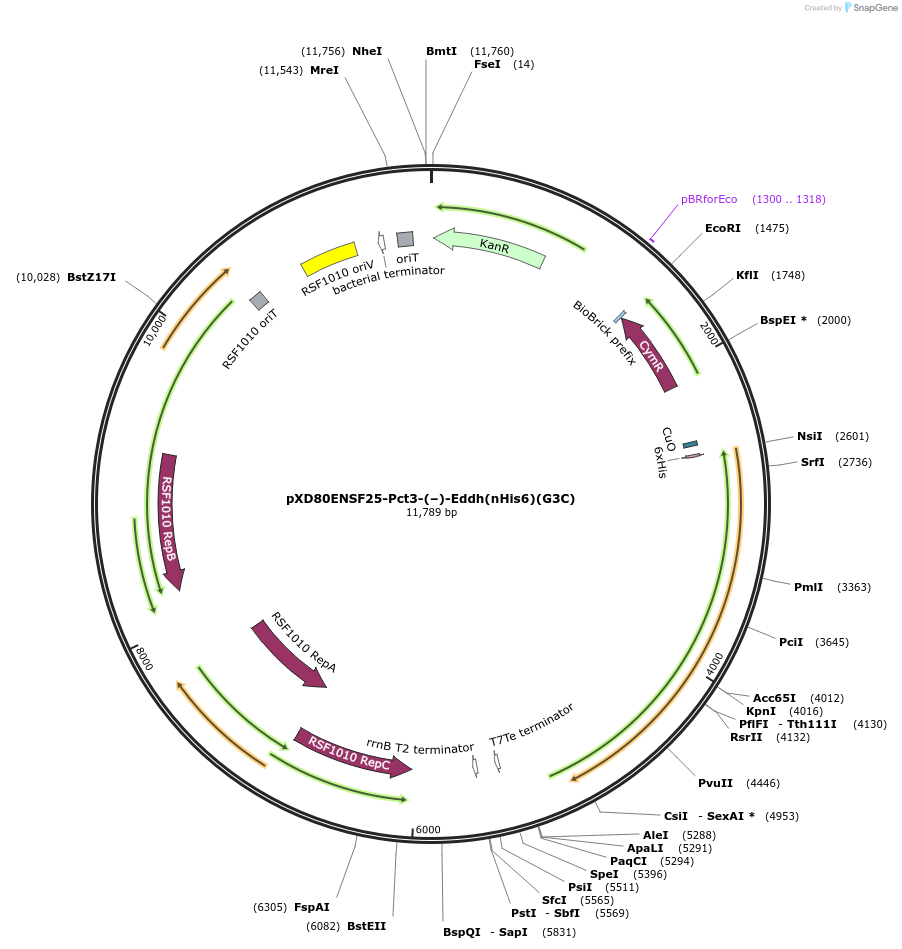
pXD80ENSF25-Pct3-(–)-Eddh(nHis6)(G3C)
Plasmid#233873PurposeExpresses His-tagged Gordonibacter pamelaeae 3C (–)-Eddh with a cumate inducible promoter in Gordonibacter urolithinfaciens DSM 27213 for protein purificationDepositorInsert(–)-Eddh
TagsHisExpressionBacterialAvailable SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
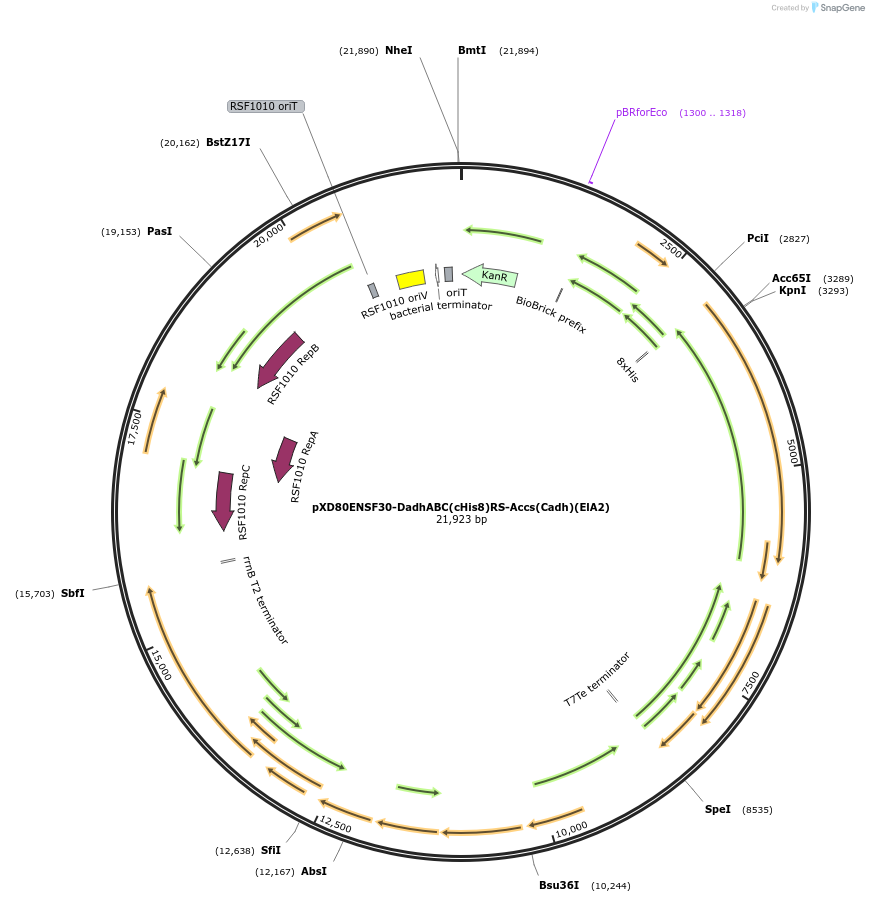
pXD80ENSF30-DadhABC(cHis8)RS-Accs(Cadh)(ElA2)
Plasmid#233876PurposeExpresses Eggerthella lenta A2 Dadh with the accessory genes from the cadh gene cluster in Gordonibacter urolithinfaciens DSM 27213DepositorInsertDadh
TagsHis8ExpressionBacterialAvailable SinceMarch 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
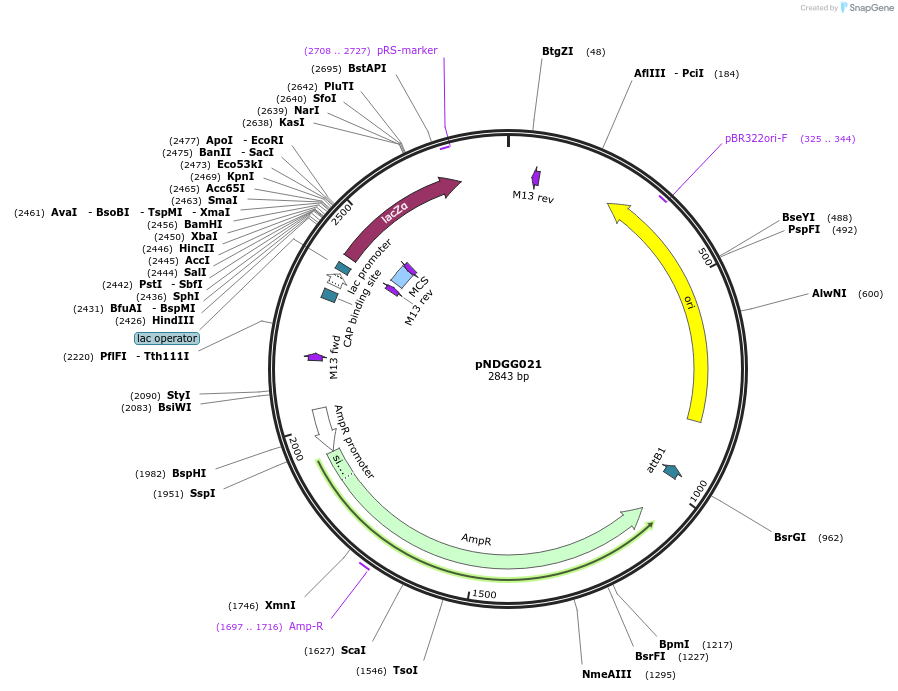
pNDGG021
Plasmid#231305PurposeVector part for BEVA; BEVA2.0 Position 1 Level 1 BsaI cloning site without T0, AmpRDepositorInsertBEVA2.0 Position 1 Level 1 BsaI cloning site without T0, AmpR
ExpressionBacterialAvailable SinceFeb. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
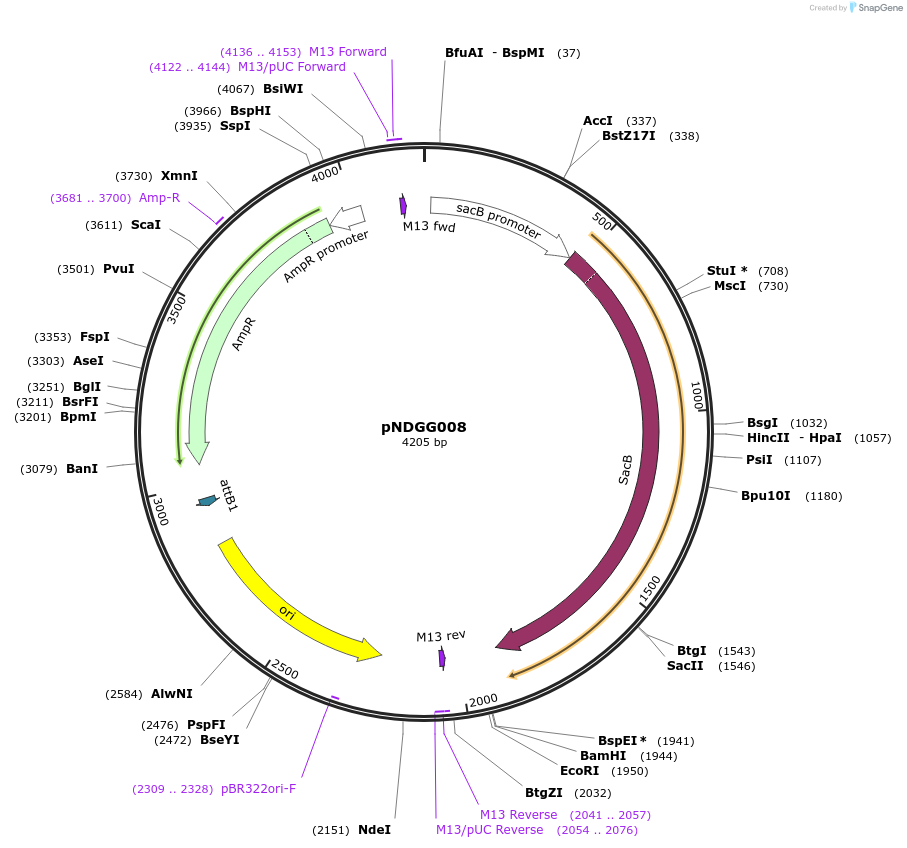
pNDGG008
Plasmid#231312PurposeVector part for BEVA; BEVA2.0 Position 4 sacB sucrose counterselection module, AmpR.DepositorInsertBEVA2.0 Position 4 sacB sucrose counterselection module, AmpR.
ExpressionBacterialAvailable SinceFeb. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
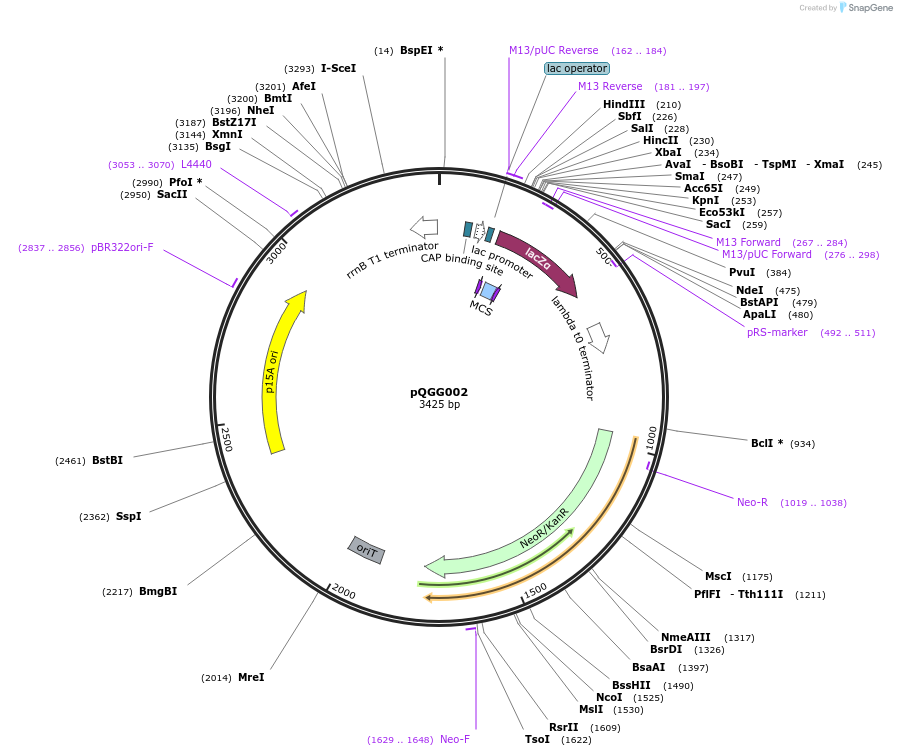
pQGG002
Plasmid#231329PurposeBEVA Golden Gate cloning vector; BEVA2.0 Narrow host range plasmid with I-SceI counterselectable site. L1-P15A-Nm-ISceI; GG assembly from pOGG004, pNDGG001, pNDGG006, pGQ0023, pOGG014. KmR/NmR.DepositorTypeEmpty backboneExpressionBacterialAvailable SinceFeb. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
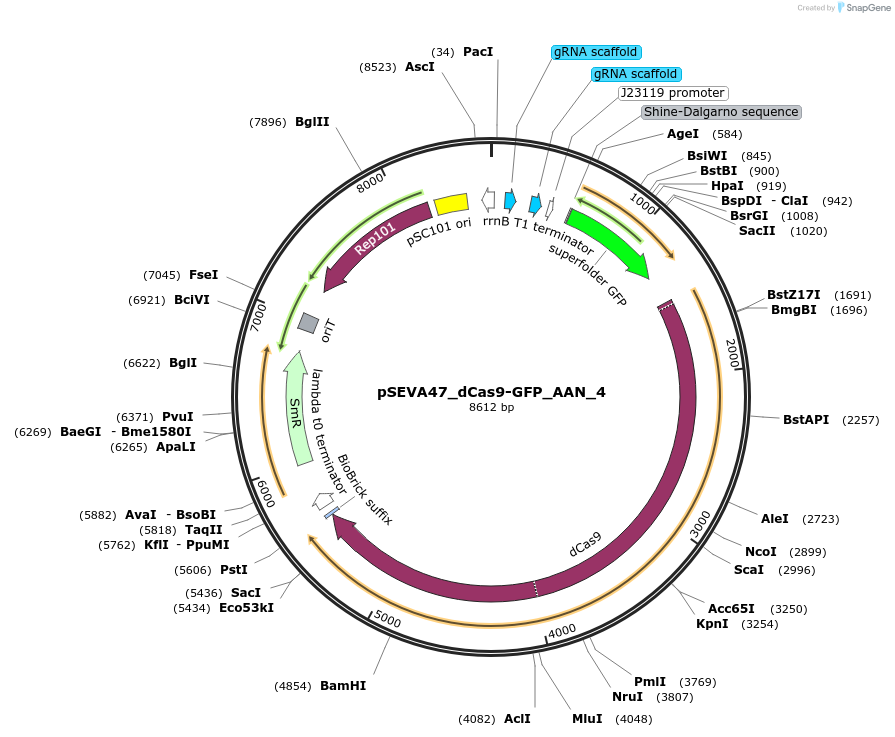
pSEVA47_dCas9-GFP_AAN_4
Plasmid#231415PurposeThis AND-AND-NOT-gate plasmid expresses dCas9 from a constitutive promoter. Its sgRNAs X & Y are repressible by sgRNAs A & B, respectively. Its GFP gene is repressible by any of sgRNAs X, Y, and C.DepositorInsertsdCas9
GFP
sgRNA-X
sgRNA-Y
UseSynthetic BiologyAvailable SinceFeb. 19, 2025AvailabilityAcademic Institutions and Nonprofits only -
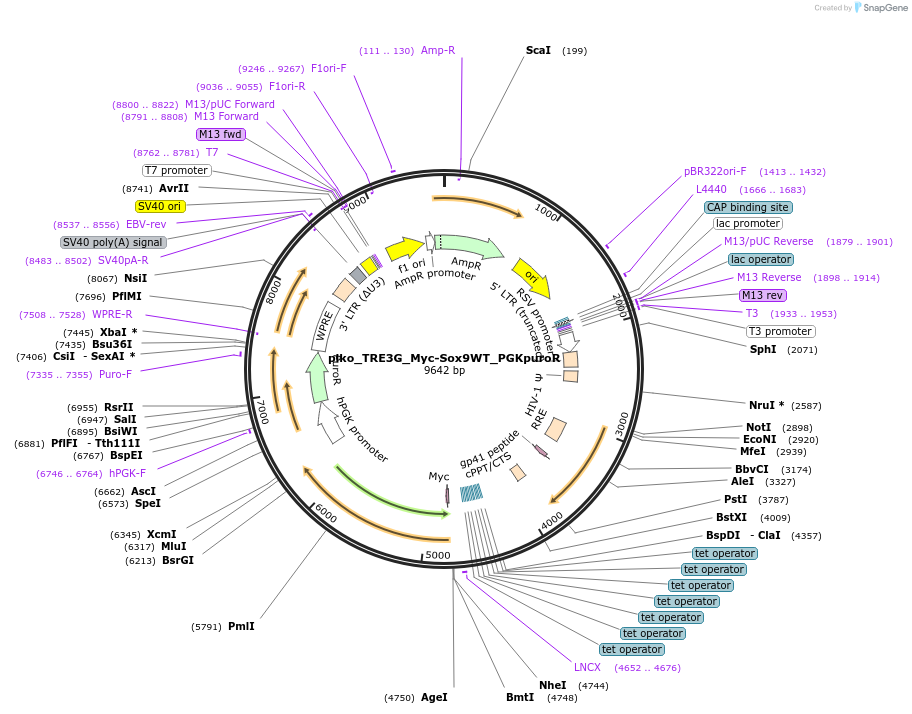
plko_TRE3G_Myc-Sox9WT_PGKpuroR
Plasmid#203962PurposeDox inducible WT SOX9 with myc tagDepositorAvailable SinceJan. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
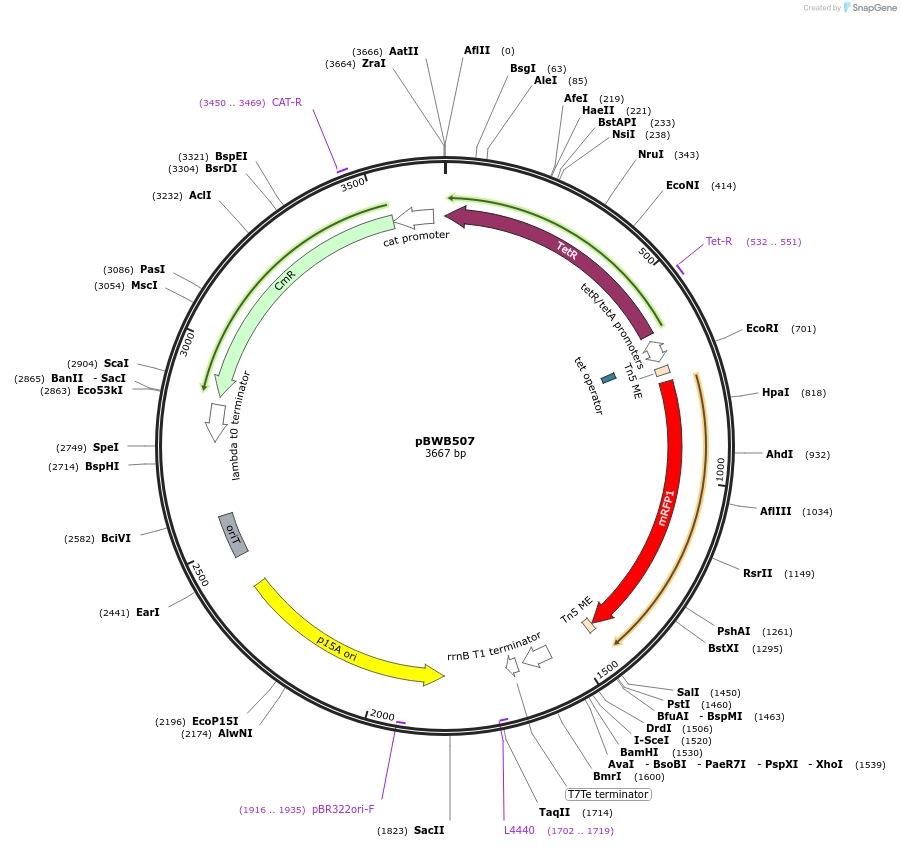
pBWB507
Plasmid#209325Purposep15A-tetR-tet-ME-RBS-mRFP-ME-U1-Barcode-U2-dbI terminator-oriT-Cm. Plasmid backbone for fragment expression.DepositorInsertmRFP
ExpressionBacterialPromotertetAvailable SinceOct. 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
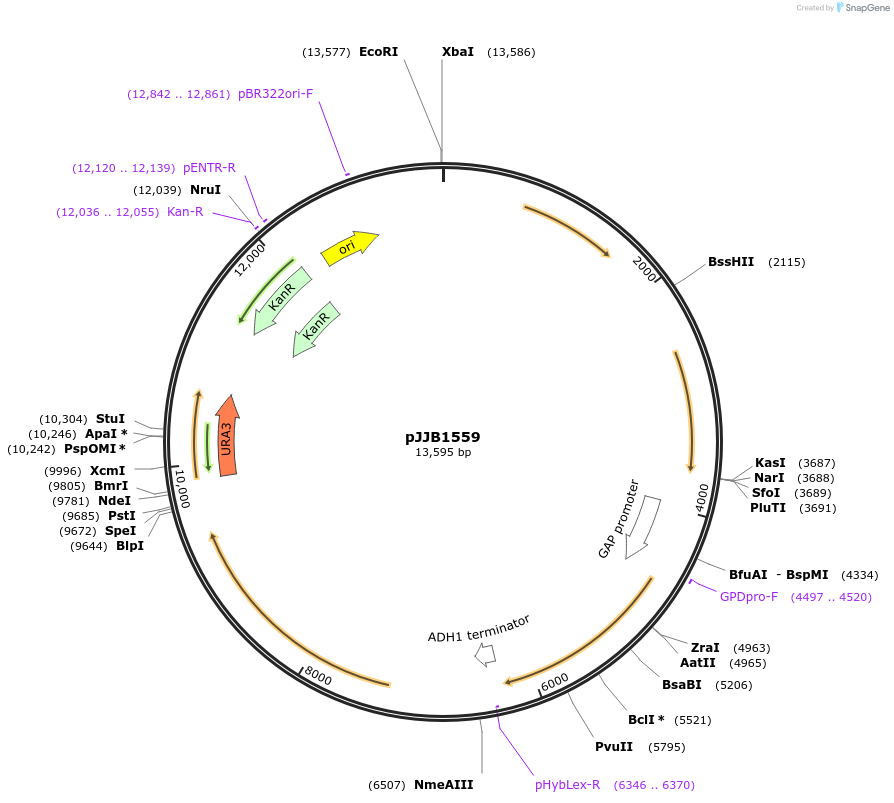
pJJB1559
Plasmid#218590PurposeExpress peroxisomal Idi1p, Erg20(F96W,N127W)p, and GESp to convert IPP to geraniol with cytosolic Erg20 homodimerDepositorInsertGES
TagsePTS1ExpressionYeastMutationErg20(F96W, N127W), Erg20-Erg20 synthetic homodim…Available SinceOct. 4, 2024AvailabilityAcademic Institutions and Nonprofits only