We narrowed to 19,614 results for: try;
-
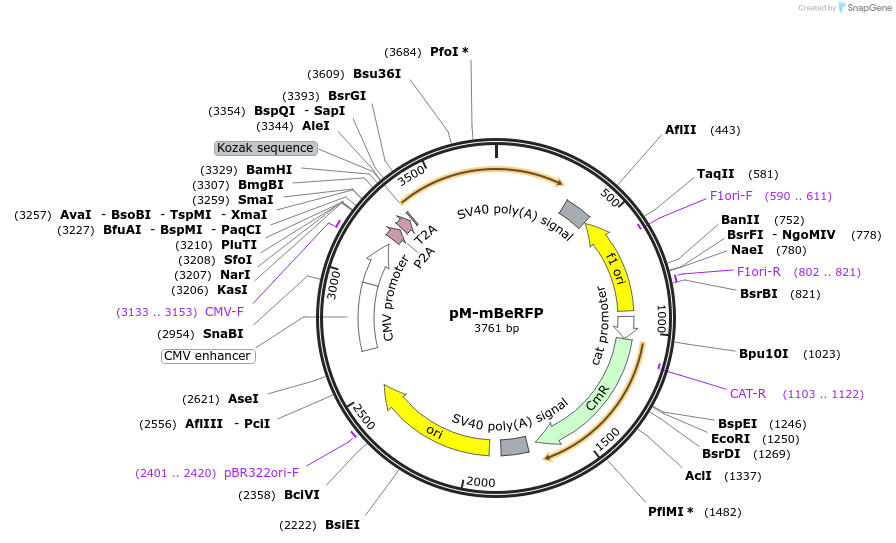
Plasmid#228620PurposeMammalian expression of mBeRFP and extra insert, for MuSIC barcode constructionDepositorInsertmBeRFP
ExpressionMammalianMutationWe introduced some silent mutations to avoid esse…PromoterCMVAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
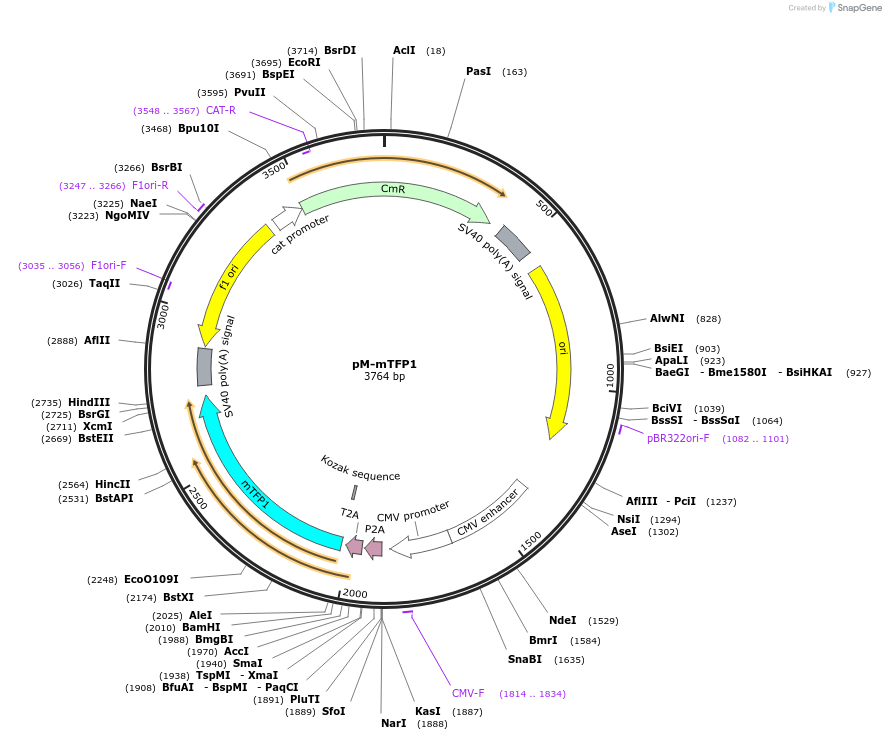
pM-mTFP1
Plasmid#228621PurposeMammalian expression of mTFP1 and extra insert, for MuSIC barcode constructionDepositorInsertmTFP1
ExpressionMammalianMutationWe introduced some silent mutations to avoid esse…PromoterCMVAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
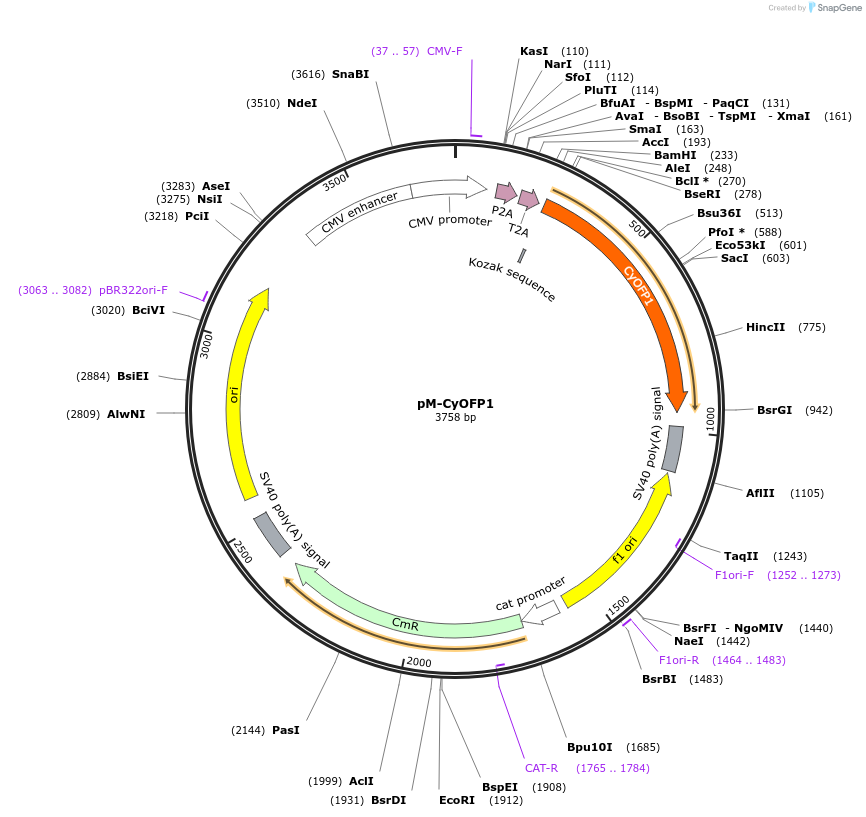
pM-CyOFP1
Plasmid#228623PurposeMammalian expression of CyOFP1 and extra insert, for MuSIC barcode constructionDepositorInsertCyOFP1
ExpressionMammalianMutationWe introduced some silent mutations to avoid esse…PromoterCMVAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
pM-mBeRFP
Plasmid#228620PurposeMammalian expression of mBeRFP and extra insert, for MuSIC barcode constructionDepositorInsertmBeRFP
ExpressionMammalianMutationWe introduced some silent mutations to avoid esse…PromoterCMVAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
pM-mTFP1
Plasmid#228621PurposeMammalian expression of mTFP1 and extra insert, for MuSIC barcode constructionDepositorInsertmTFP1
ExpressionMammalianMutationWe introduced some silent mutations to avoid esse…PromoterCMVAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
pM-CyOFP1
Plasmid#228623PurposeMammalian expression of CyOFP1 and extra insert, for MuSIC barcode constructionDepositorInsertCyOFP1
ExpressionMammalianMutationWe introduced some silent mutations to avoid esse…PromoterCMVAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
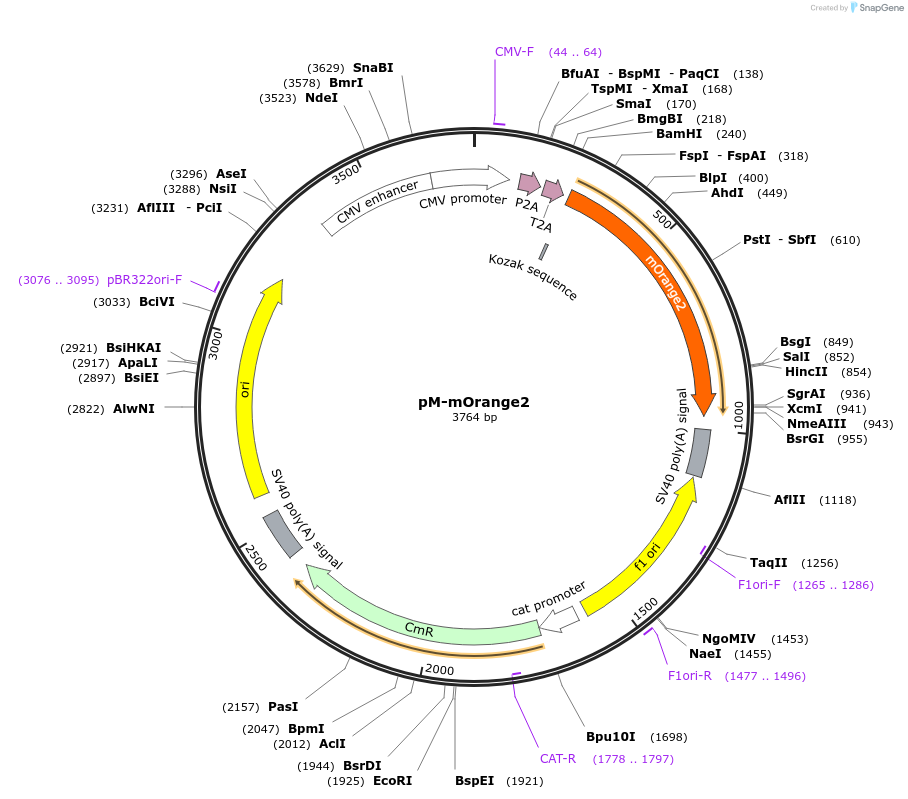
pM-mOrange2
Plasmid#228627PurposeMammalian expression of mOrange2 and extra insert, for MuSIC barcode constructionDepositorInsertmOrange2
ExpressionMammalianMutationWe introduced some silent mutations to avoid esse…PromoterCMVAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
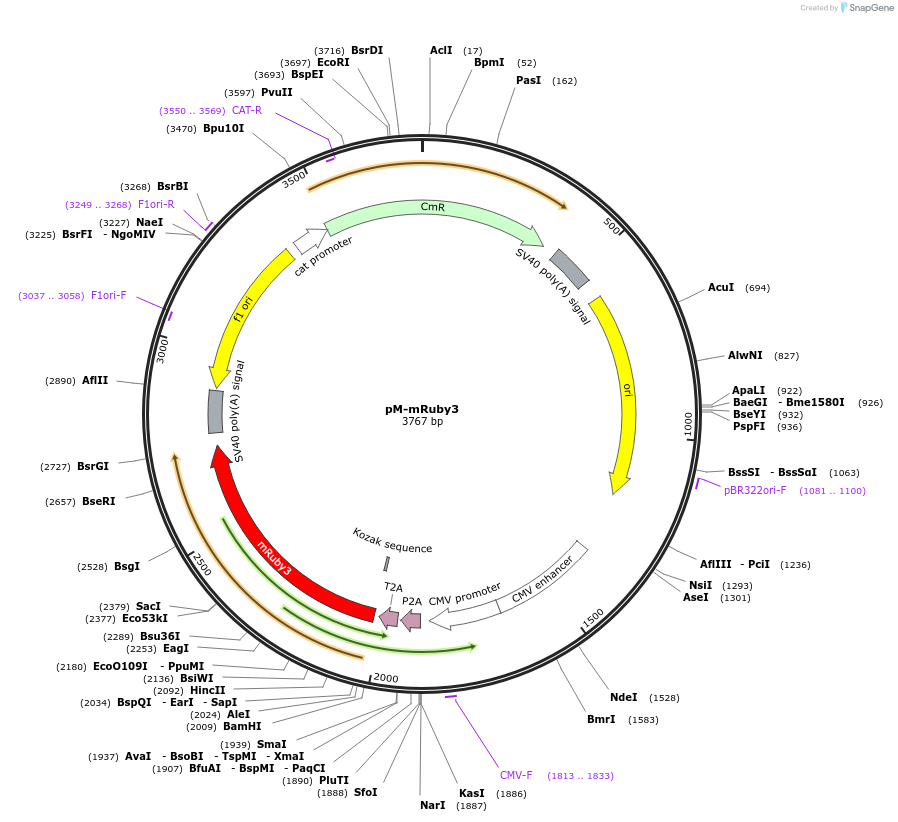
pM-mRuby3
Plasmid#228628PurposeMammalian expression of mRuby3 and extra insert, for MuSIC barcode constructionDepositorInsertmRuby3
ExpressionMammalianMutationWe introduced some silent mutations to avoid esse…PromoterCMVAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
pM-mOrange2
Plasmid#228627PurposeMammalian expression of mOrange2 and extra insert, for MuSIC barcode constructionDepositorInsertmOrange2
ExpressionMammalianMutationWe introduced some silent mutations to avoid esse…PromoterCMVAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
pM-mRuby3
Plasmid#228628PurposeMammalian expression of mRuby3 and extra insert, for MuSIC barcode constructionDepositorInsertmRuby3
ExpressionMammalianMutationWe introduced some silent mutations to avoid esse…PromoterCMVAvailable SinceNov. 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
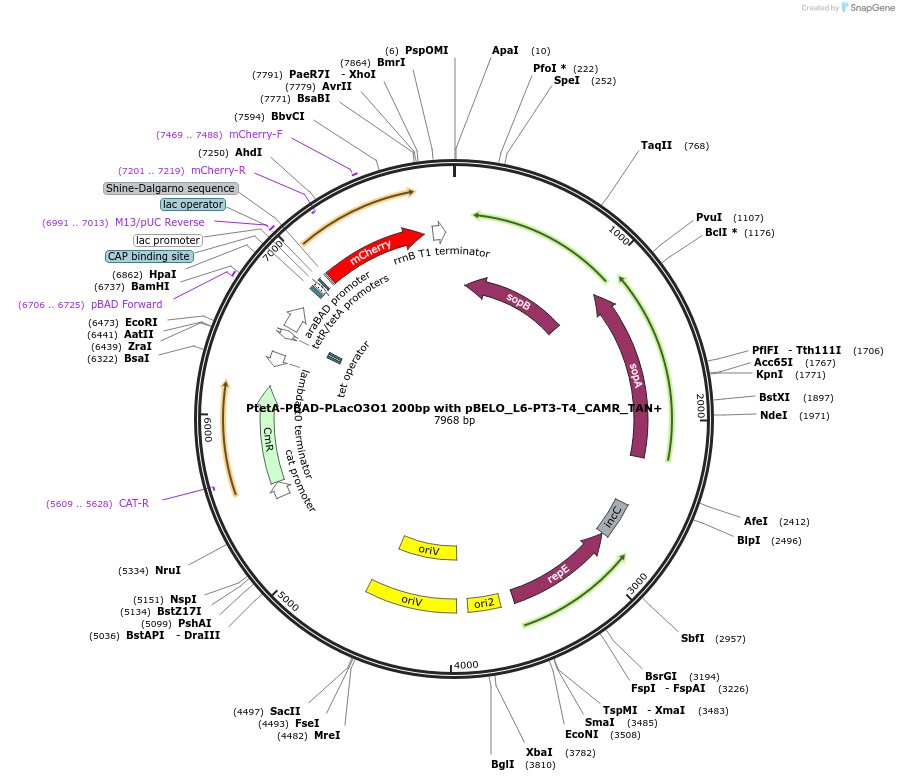
PtetA-PBAD-PLacO3O1 200bp with pBELO_L6-PT3-T4_CAMR_TAN+
Plasmid#225649PurposetetA, araBAD and LacO3O1 promoters in tandem conformation with 200bp distance between them, expressing mCherry with pBELO_L6-PT3-T4 backbone and chloramphenicol resistanceDepositorInserttetR/tetA, araBAD and LacO3O1 in tandem conformation
TagsmCherryExpressionBacterialPromotertetA, araBAD and LacO3O1 promotersAvailable SinceOct. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
PtetA-PBAD-PLacO3O1 200bp with pBELO_L6-PT3-T4_CAMR_TAN+
Plasmid#225649PurposetetA, araBAD and LacO3O1 promoters in tandem conformation with 200bp distance between them, expressing mCherry with pBELO_L6-PT3-T4 backbone and chloramphenicol resistanceDepositorInserttetR/tetA, araBAD and LacO3O1 in tandem conformation
TagsmCherryExpressionBacterialPromotertetA, araBAD and LacO3O1 promotersAvailable SinceOct. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
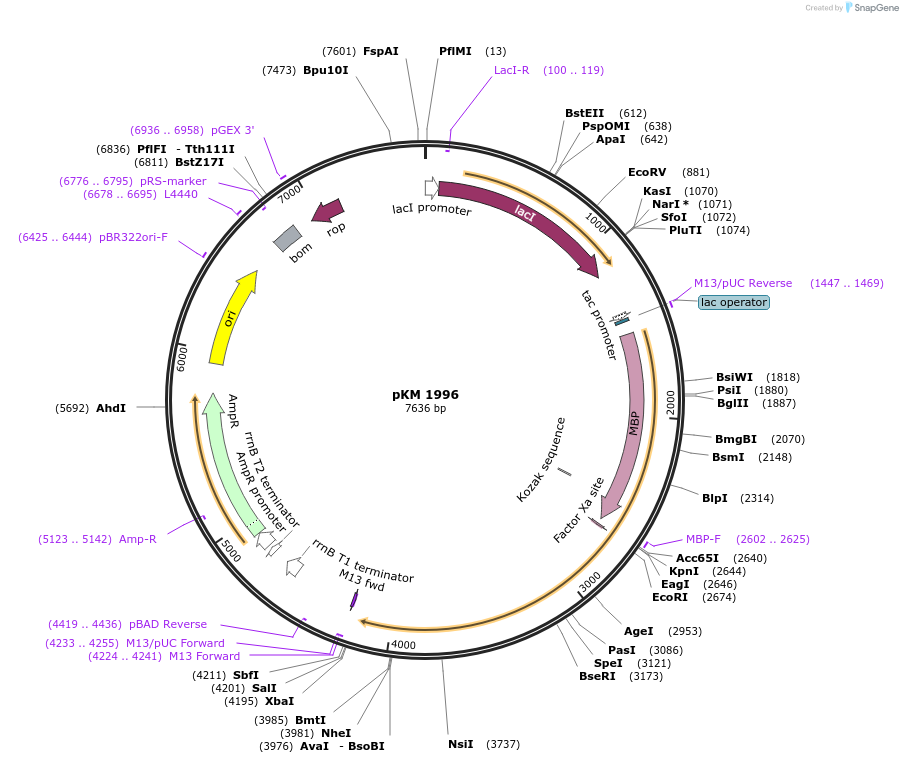
pKM 1996
Plasmid#217825PurposeExpresses putative photoreceptor SA-phr1 from S. alba. This gene shows high homology to the phr genes in prokaryotes.DepositorInsertSA-phr1
TagsMaltose-binding proteinExpressionBacterialAvailable SinceJune 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
pKM 1996
Plasmid#217825PurposeExpresses putative photoreceptor SA-phr1 from S. alba. This gene shows high homology to the phr genes in prokaryotes.DepositorInsertSA-phr1
TagsMaltose-binding proteinExpressionBacterialAvailable SinceJune 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
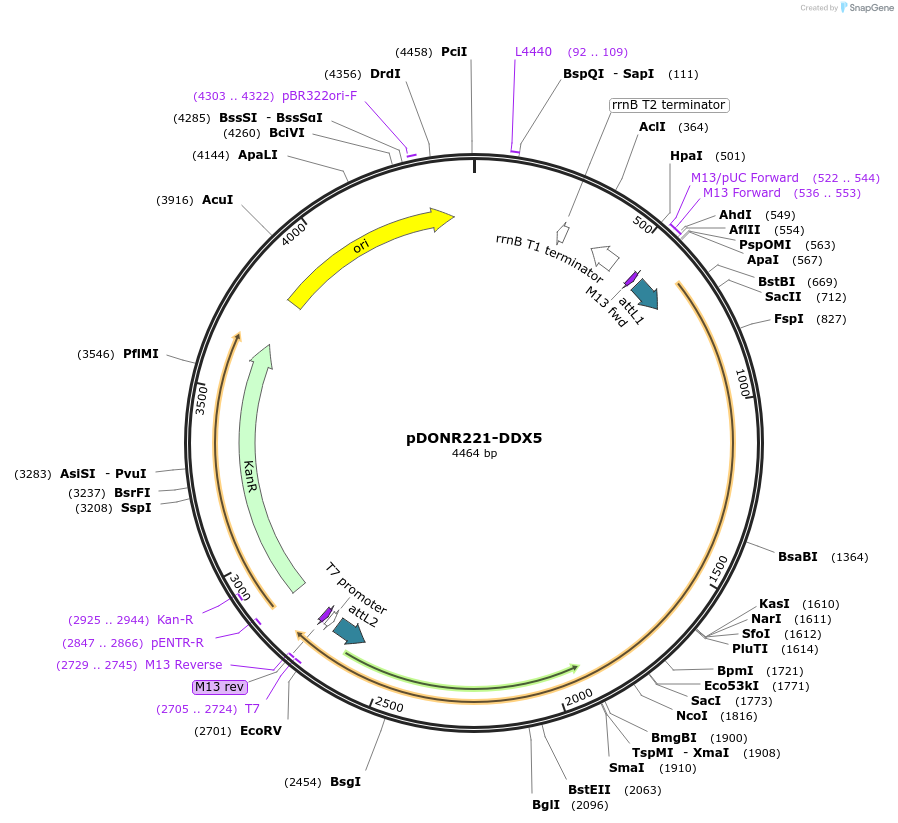
pDONR221-DDX5
Plasmid#166163PurposeGateway entry vector for N. furzeriDDX5DepositorInsertDDX5
UseGateway entry vectorAvailable SinceMay 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
pDONR221-DDX5
Plasmid#166163PurposeGateway entry vector for N. furzeriDDX5DepositorInsertDDX5
UseGateway entry vectorAvailable SinceMay 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
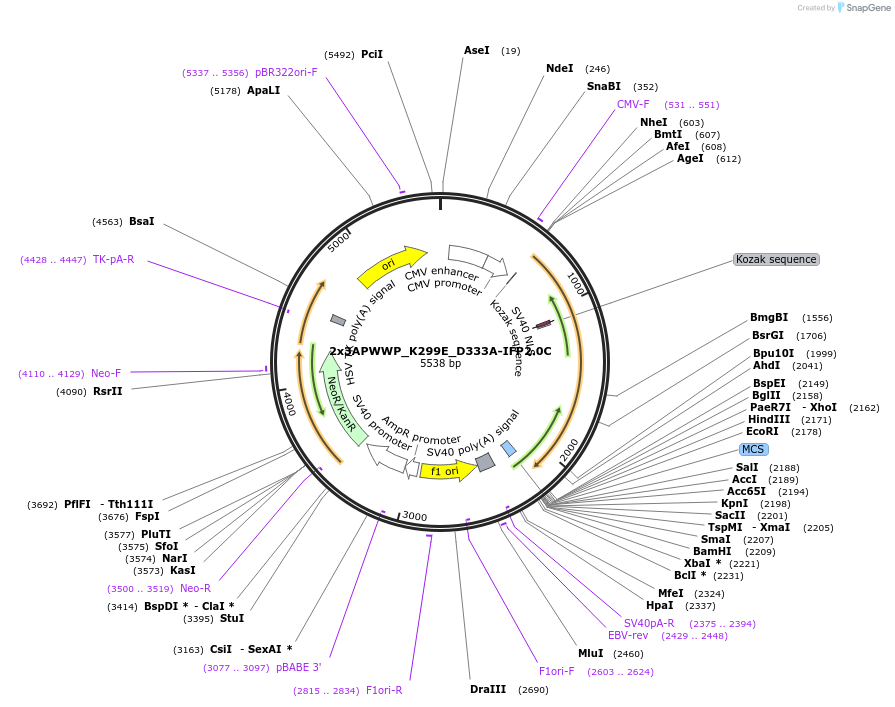
2x3APWWP_K299E_D333A-IFP2.0C
Plasmid#215753PurposeExpresses binding-deficient K299E/D333A mutant of 2xDNMT3A PWWP domain fused to C-terminal part of IFP2.0 as negative control for H3K36me2/3 detection in BiAD sensorsDepositorInsert2x3APWWP_K299E_D333A-IFP2.0C
ExpressionMammalianPromoterCMVAvailable SinceApril 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
2x3APWWP_K299E_D333A-IFP2.0C
Plasmid#215753PurposeExpresses binding-deficient K299E/D333A mutant of 2xDNMT3A PWWP domain fused to C-terminal part of IFP2.0 as negative control for H3K36me2/3 detection in BiAD sensorsDepositorInsert2x3APWWP_K299E_D333A-IFP2.0C
ExpressionMammalianPromoterCMVAvailable SinceApril 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
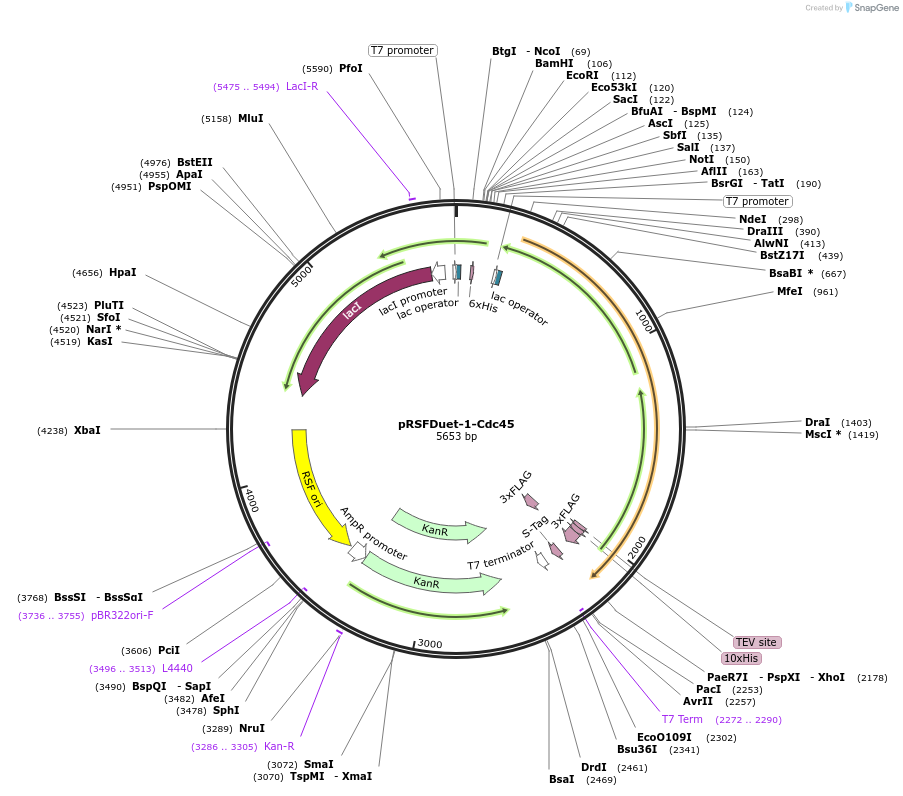
pRSFDuet-1-Cdc45
Plasmid#208796PurposeXenopus laevis FLAG5-His10-TEV-Cdc45DepositorAvailable SinceFeb. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
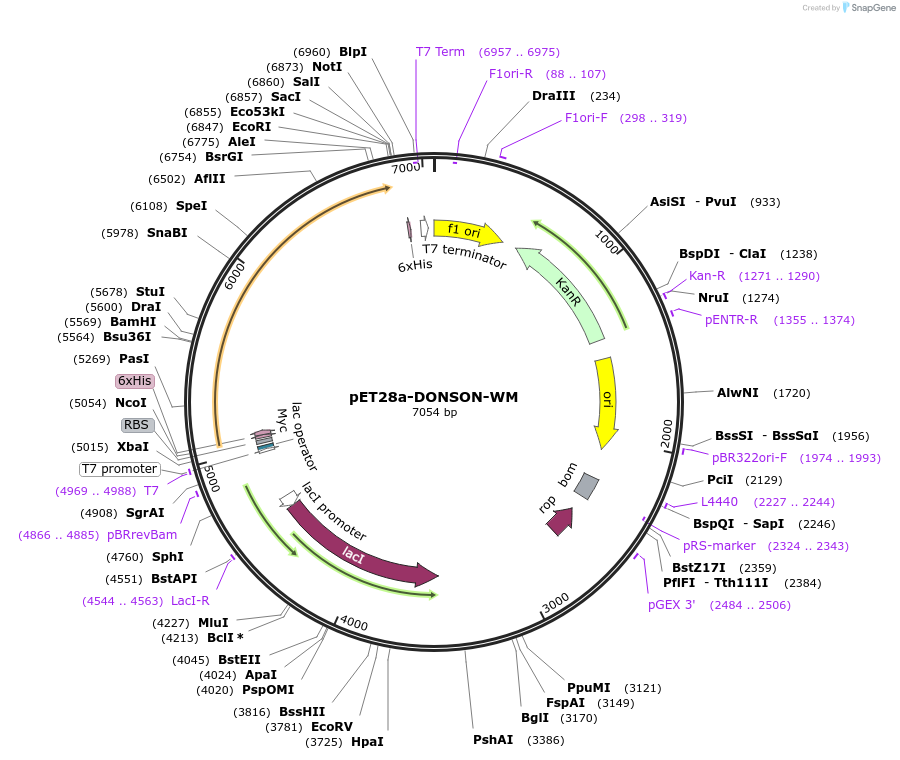
pET28a-DONSON-WM
Plasmid#208798PurposeXenopus laevis His6-Myc-DONSON (W234L, M463T)DepositorInsertDONSON (donson.L Frog)
TagsHis and MycExpressionBacterialMutationW234L; M463TPromoterT7Available SinceFeb. 8, 2024AvailabilityAcademic Institutions and Nonprofits only