We narrowed to 8,112 results for: chloramphenicol
-
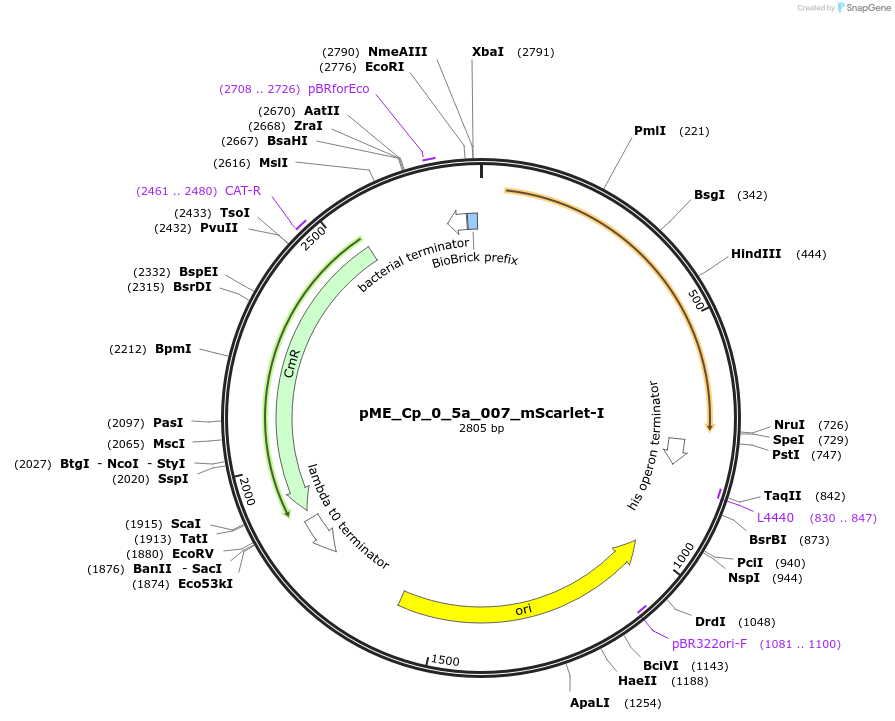
Plasmid#235889PurposemScarlet-I C-tagDepositorInsertmScarlet-I C-tag
UseSynthetic BiologyAvailable SinceJune 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
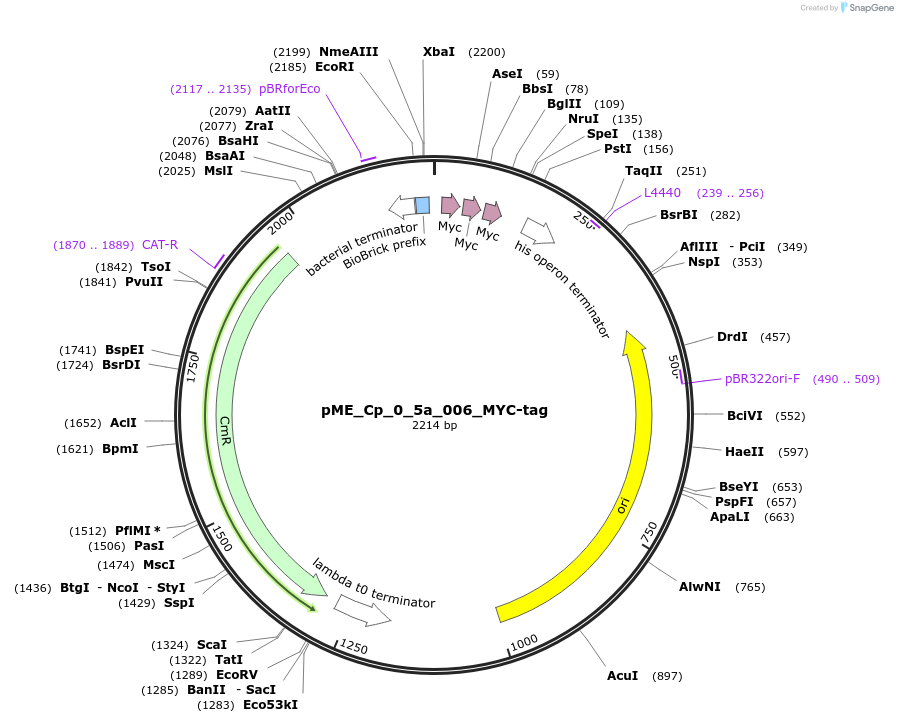
pME_Cp_0_5a_006_MYC-tag
Plasmid#235888PurposeMYC C-tagDepositorInsertMYC C-tag
UseSynthetic BiologyAvailable SinceJune 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
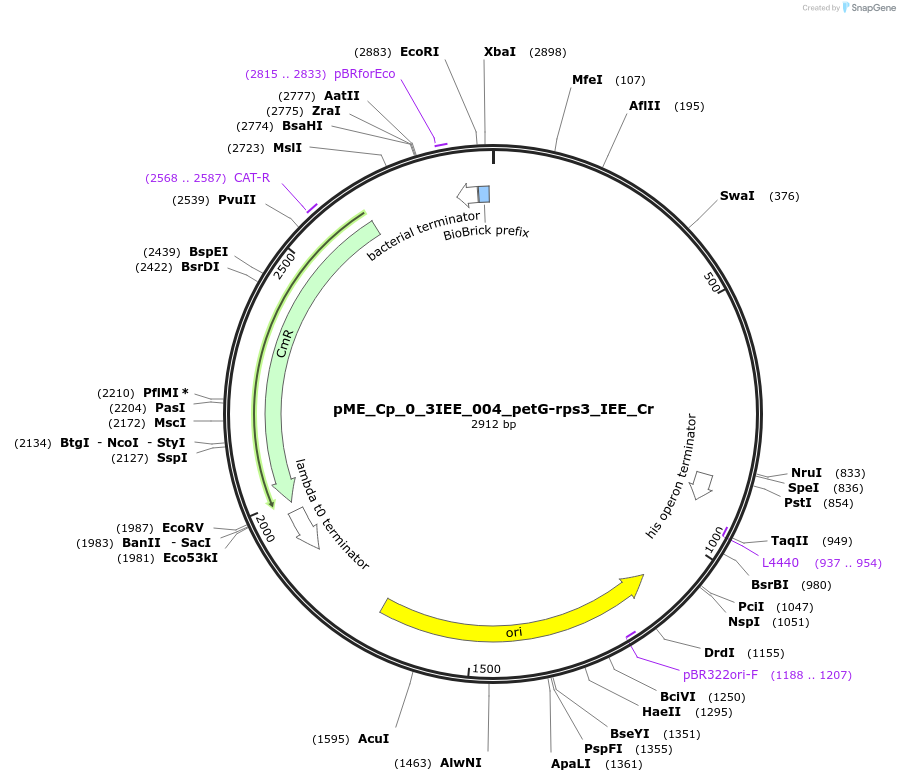
pME_Cp_0_3IEE_004_petG-rps3_IEE_Cr
Plasmid#235861PurposepetG-rps3 IEE Chlamydomonas reinhardtiiDepositorInsertpetG-rps3 IEE
UseSynthetic BiologyMutationnoneAvailable SinceJune 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
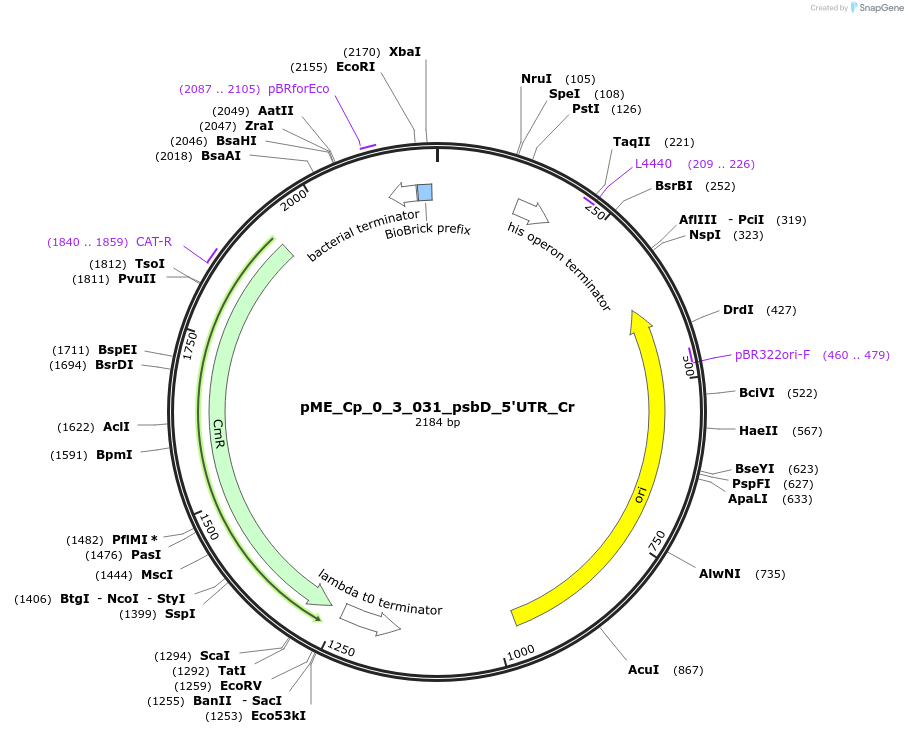
pME_Cp_0_3_031_psbD_5'UTR_Cr
Plasmid#235850PurposepsbD 5'UTR Chlamydomonas reinhardtiiDepositorInsertpsbD 5'UTR
UseSynthetic BiologyMutationnoneAvailable SinceJune 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
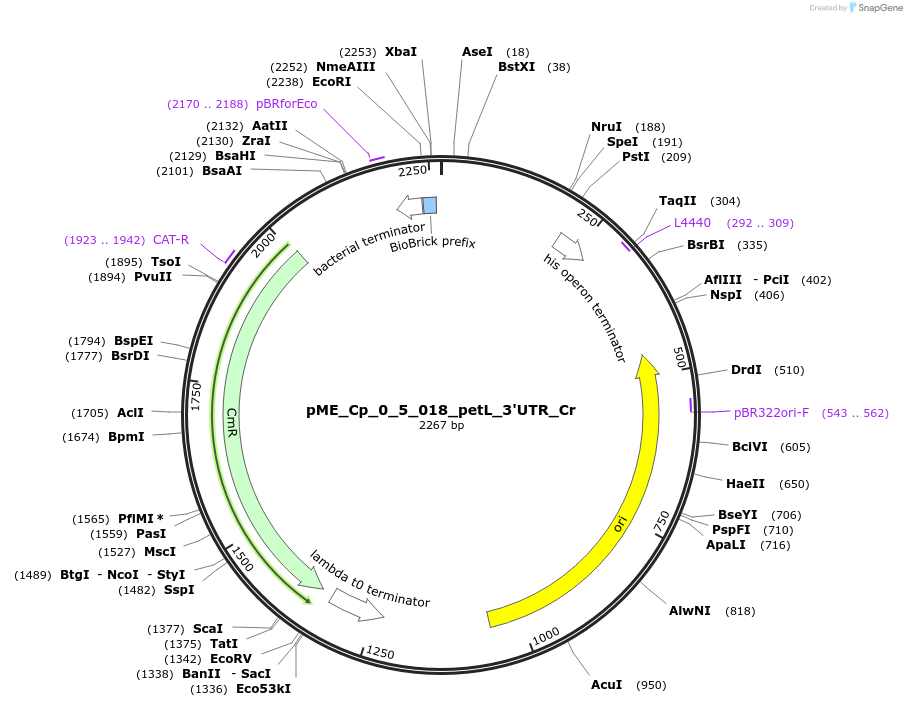
pME_Cp_0_5_018_petL_3'UTR_Cr
Plasmid#235916PurposepetL 3'UTR Chlamydomonas reinhardtiiDepositorInsertpetL 3'UTR
UseSynthetic BiologyAvailable SinceJune 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
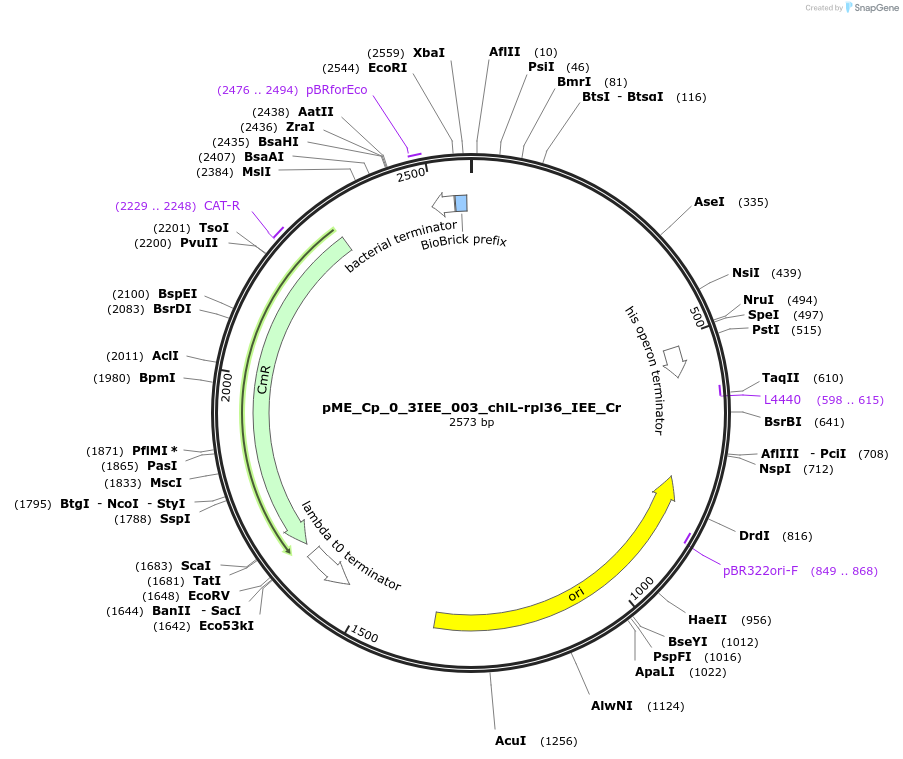
pME_Cp_0_3IEE_003_chlL-rpl36_IEE_Cr
Plasmid#235860PurposechlL-rpl36 IEE Chlamydomonas reinhardtiiDepositorInsertchlL-rpl36 IEE
UseSynthetic BiologyMutationnoneAvailable SinceJune 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
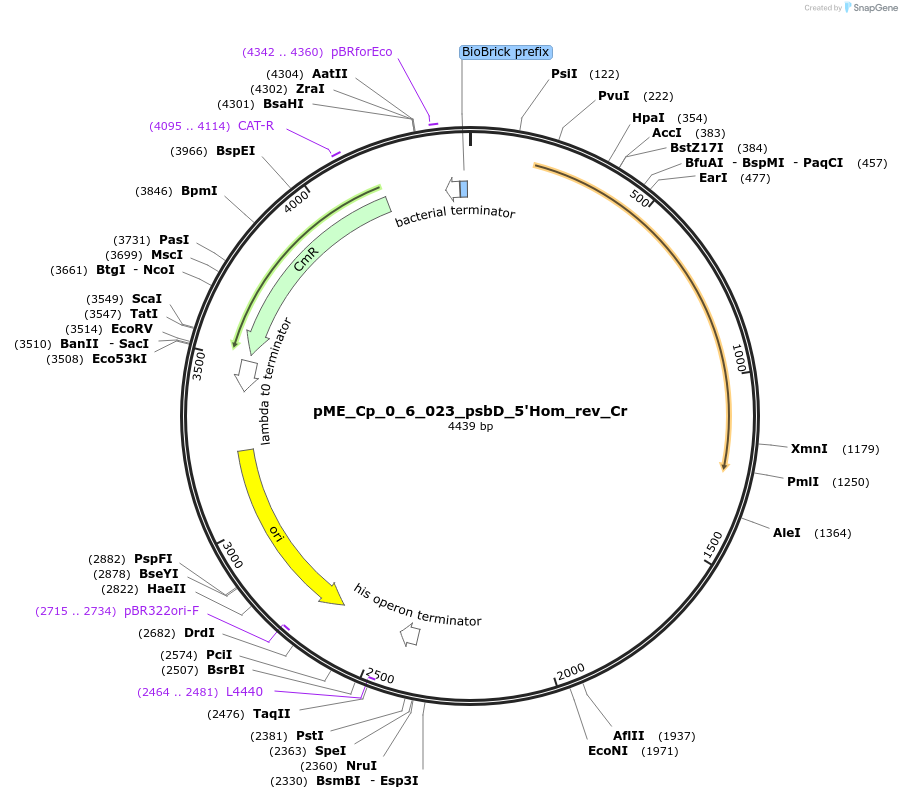
pME_Cp_0_6_023_psbD_5'Hom_rev_Cr
Plasmid#235948Purpose5'Homology psbD reverse in the M2 position used for genome integration at the psbD locusDepositorInsertpsbD 5'Homology reverse
UseSynthetic BiologyAvailable SinceJune 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
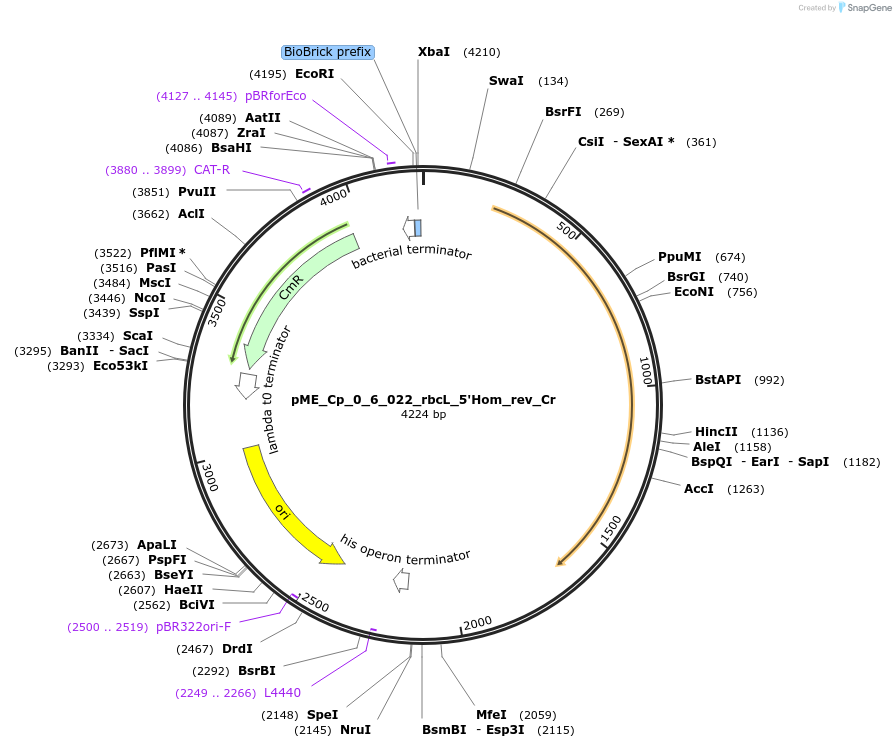
pME_Cp_0_6_022_rbcL_5'Hom_rev_Cr
Plasmid#235947Purpose5'Homology rbcL reverse in the M2 position used for genome integration at the rbcL locusDepositorInsertrbcL 5'Homology reverse
UseSynthetic BiologyAvailable SinceJune 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
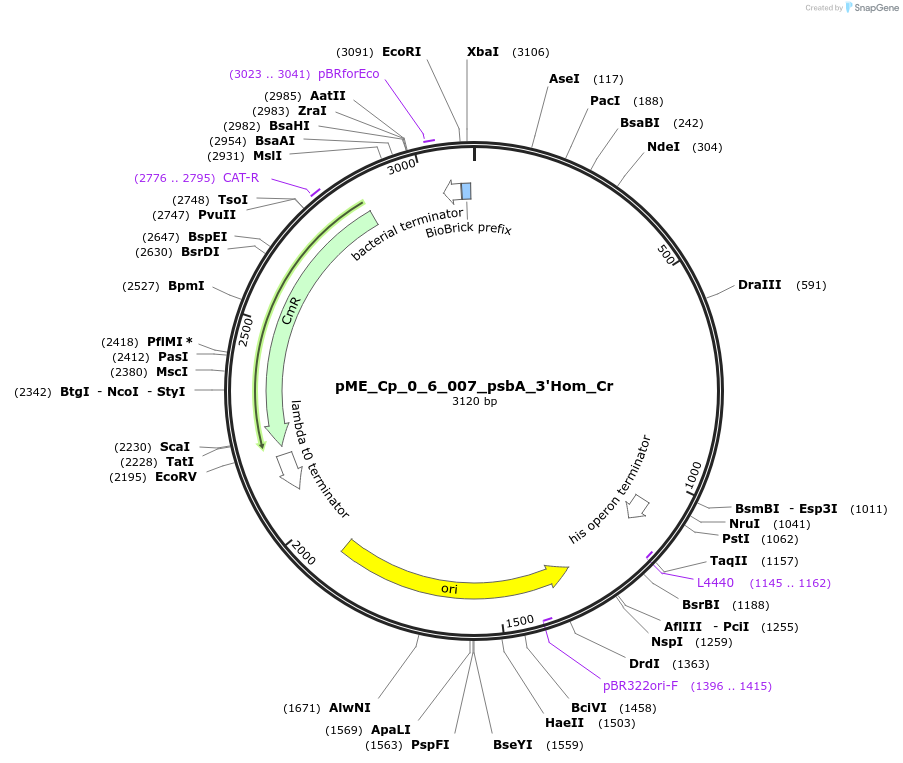
pME_Cp_0_6_007_psbA_3'Hom_Cr
Plasmid#235941Purpose3' Homology psbA in the M2 position used for genome integration at the psbA locusDepositorInsertpsbA 3'Homology
UseSynthetic BiologyAvailable SinceJune 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
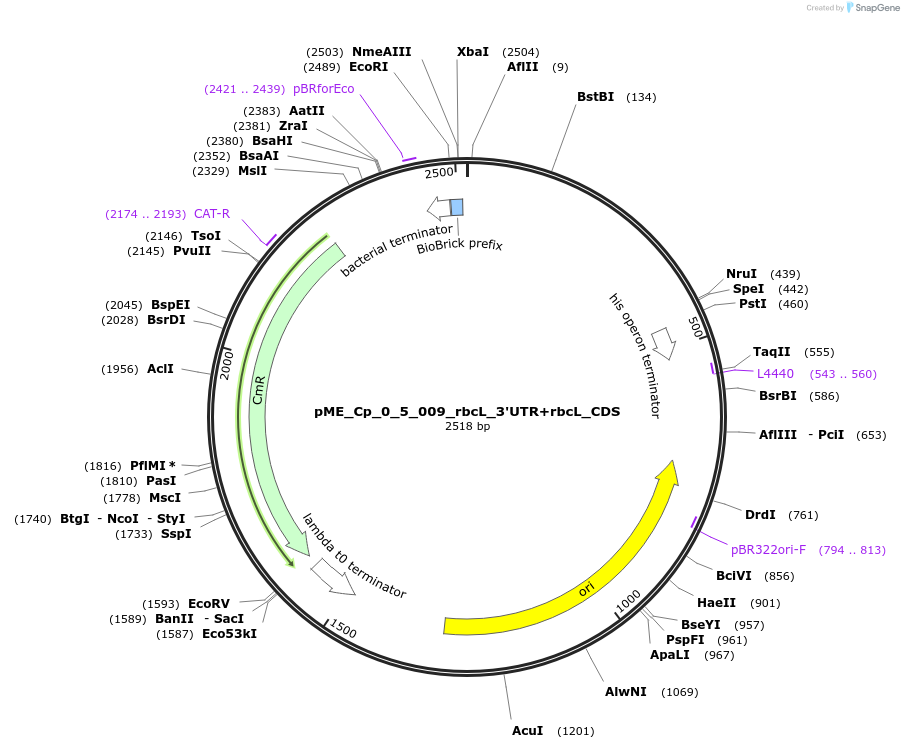
pME_Cp_0_5_009_rbcL_3'UTR+rbcL_CDS
Plasmid#235911PurposerbcL 3'UTR + rbcL CDS Chlamydomonas reinhardtiiDepositorInsertrbcL 3'UTR
UseSynthetic BiologyAvailable SinceJune 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
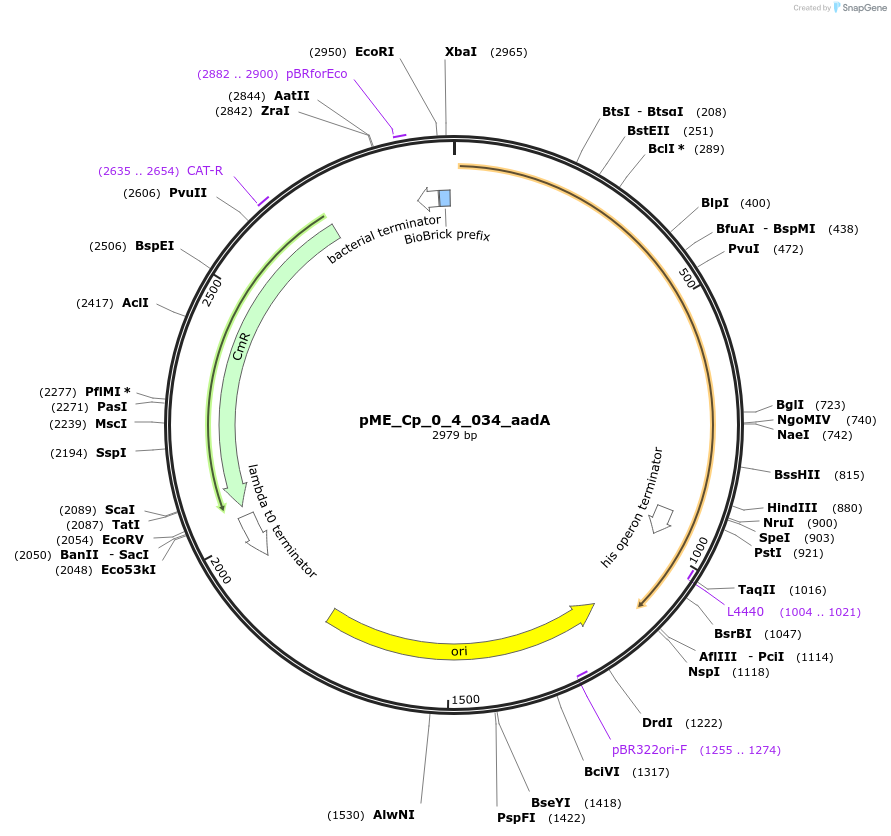
pME_Cp_0_4_034_aadA
Plasmid#235903PurposeAminoglycoside-3''-9-adenylyltransferase selection gene, codon optimized for Chlamydomonas reinhardtiiDepositorInsertaadA
UseSynthetic BiologyAvailable SinceJune 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
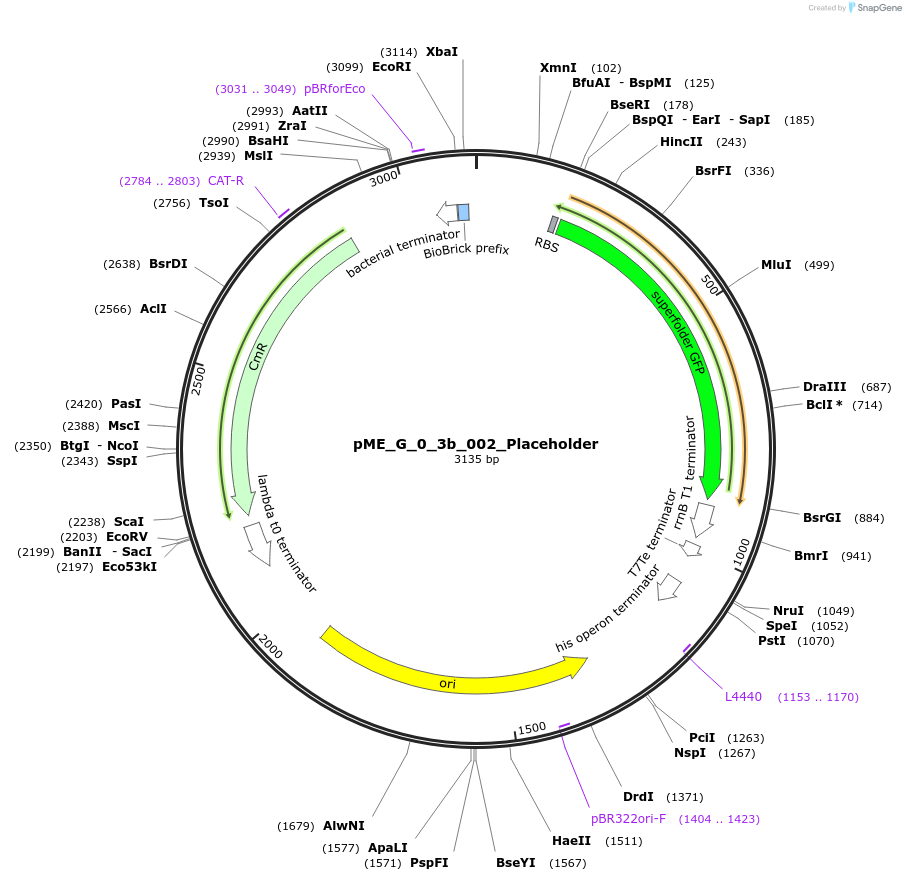
pME_G_0_3b_002_Placeholder
Plasmid#235877PurposeEncodes a sfGFP cassette in the N-tag position used for placeholder cloningDepositorInsertN-tag placeholder
UseSynthetic BiologyAvailable SinceJune 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
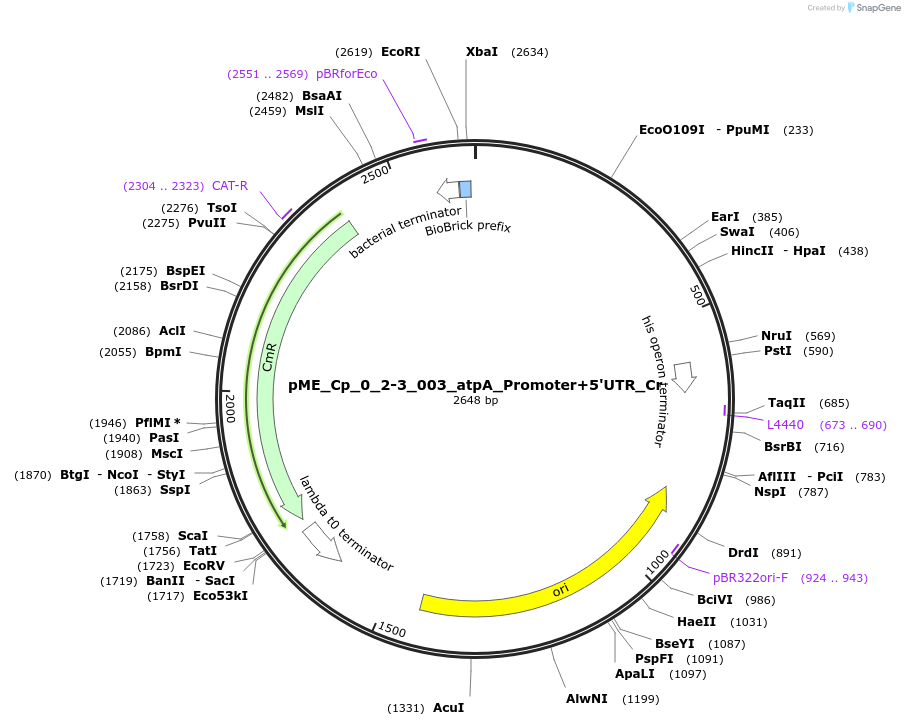
pME_Cp_0_2-3_003_atpA_Promoter+5'UTR_Cr
Plasmid#235873PurposeatpA promoter + 5'UTR Chlamydomonas reinhardtiiDepositorInsertatpA promoter + 5'UTR
UseSynthetic BiologyAvailable SinceJune 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
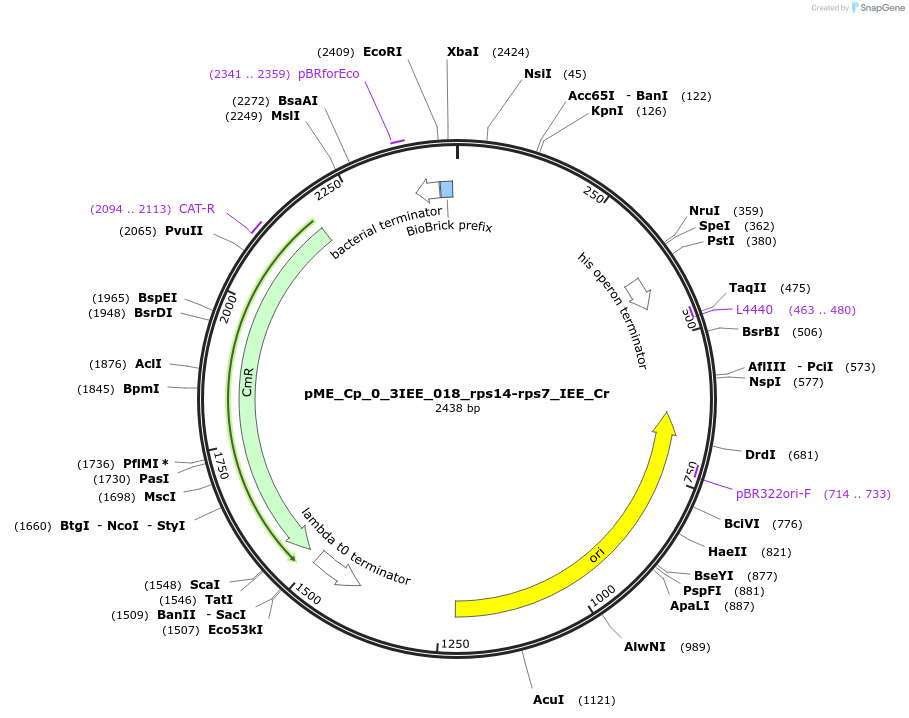
pME_Cp_0_3IEE_018_rps14-rps7_IEE_Cr
Plasmid#235866Purposerps14-rps7 IEE Chlamydomonas reinhardtiiDepositorInsertrps14-rps7 IEE
UseSynthetic BiologyMutationnoneAvailable SinceJune 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
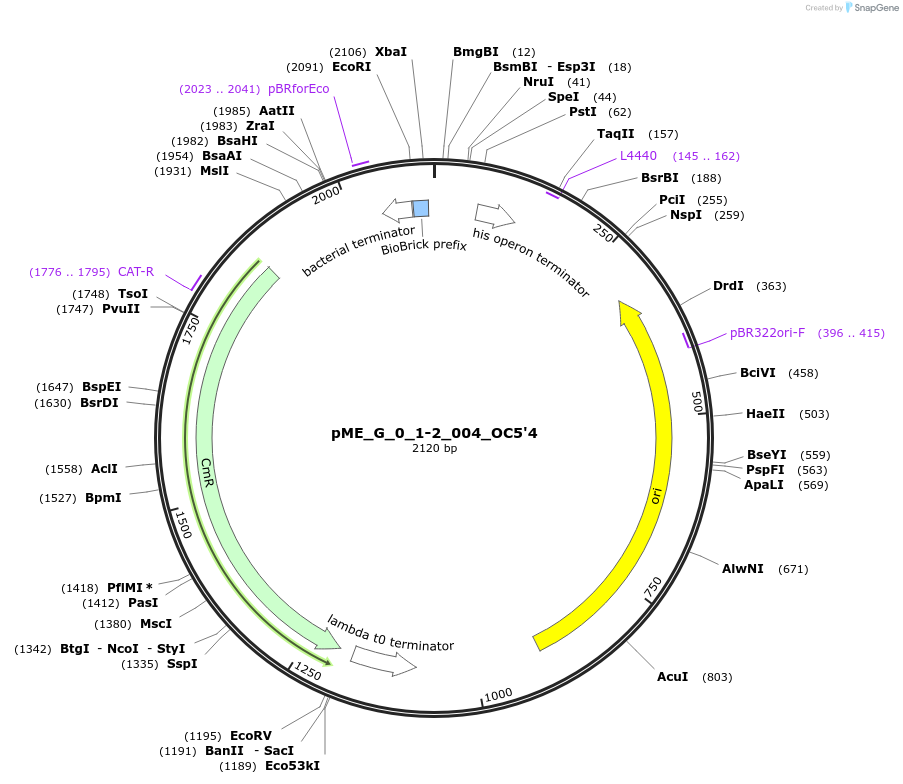
pME_G_0_1-2_004_OC5'4
Plasmid#235804Purpose5' Operon Connector 4 in the M1 position used to construct operonsDepositorInsert5' Operon Connector 4
UseSynthetic BiologyMutationnoneAvailable SinceJune 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
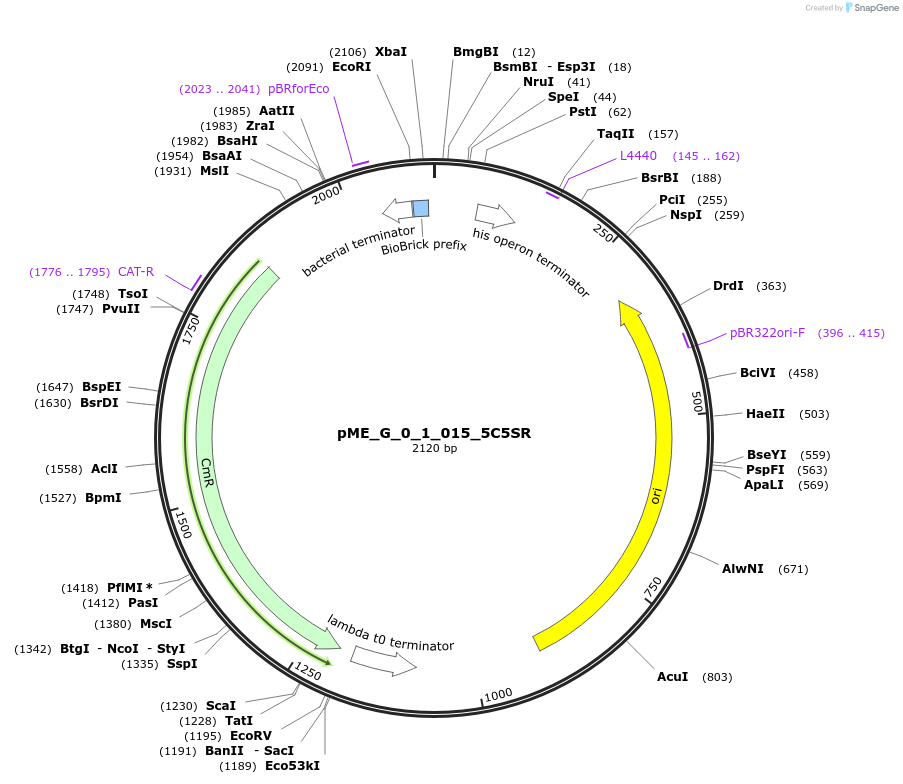
pME_G_0_1_015_5C5SR
Plasmid#235799Purpose5' Connector 6 Short Reverse in the M1 position used to dictate position for lvl2 assemblyDepositorInsert5' Connector 6 Short Reverse
UseSynthetic BiologyMutationnoneAvailable SinceJune 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
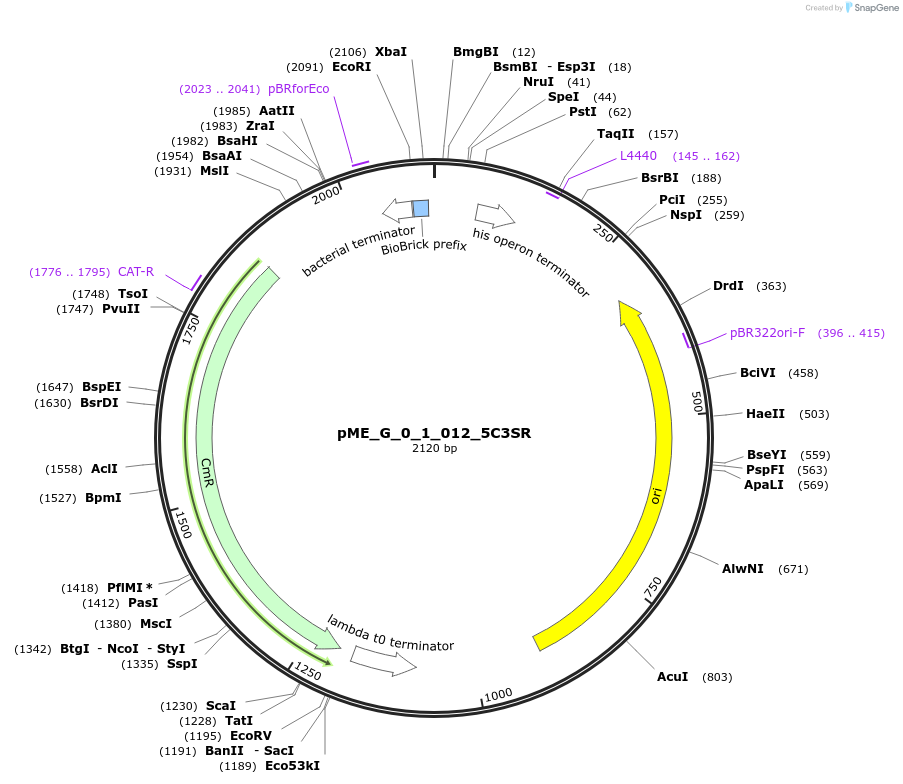
pME_G_0_1_012_5C3SR
Plasmid#235798Purpose5' Connector 3 Short Reverse in the M1 position used to dictate position for lvl2 assemblyDepositorInsert5' Connector 3 Short Reverse
UseSynthetic BiologyMutationnoneAvailable SinceJune 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
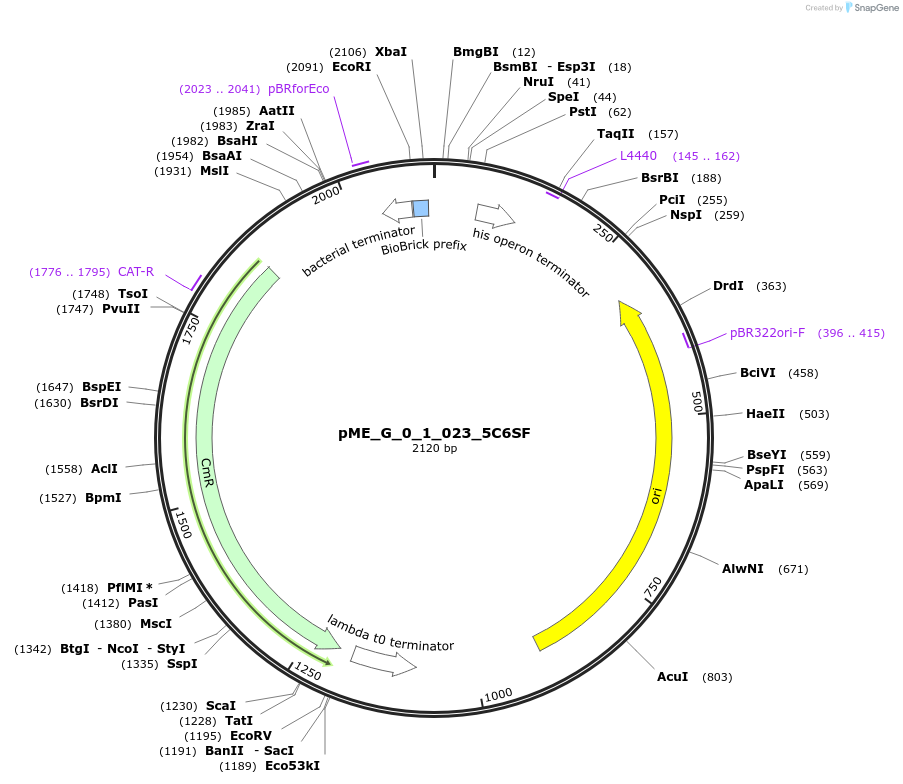
pME_G_0_1_023_5C6SF
Plasmid#235794Purpose5' Connector 6 Short Forward in the M1 position used to dictate position for lvl2 assemblyDepositorInsert5' Connector 6 Short Forward
UseSynthetic BiologyMutationnoneAvailable SinceJune 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
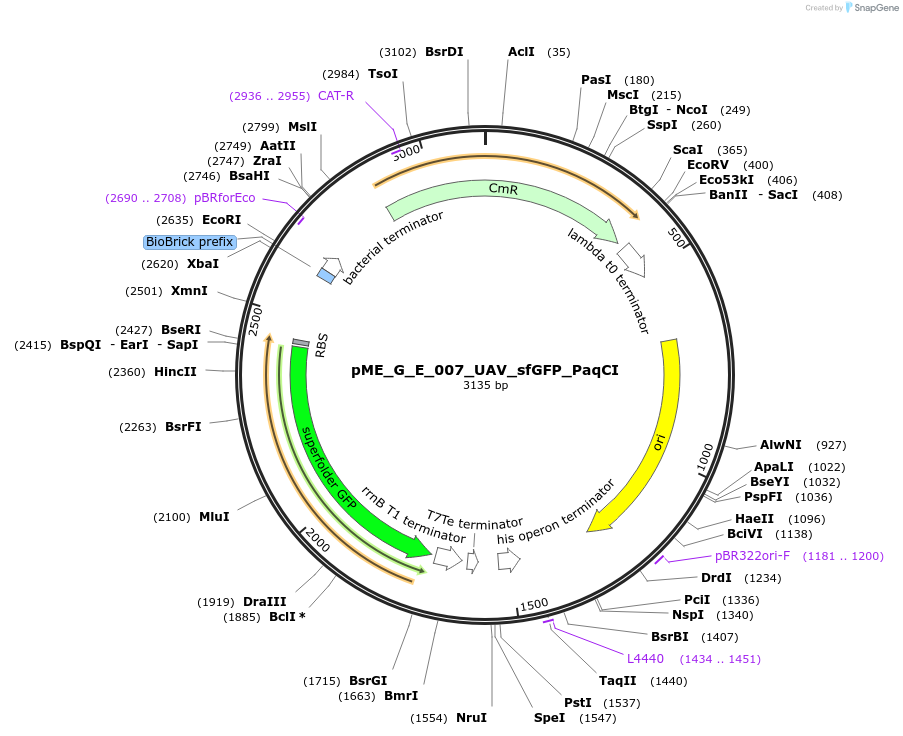
pME_G_E_007_UAV_sfGFP_PaqCI
Plasmid#235975PurposeUniversal acceptor vector with sfGFP placeholder PaqCI compatible for lvl0 assemblyDepositorInsertUAV_sfGFP_PaqCI
UseSynthetic BiologyAvailable SinceMay 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
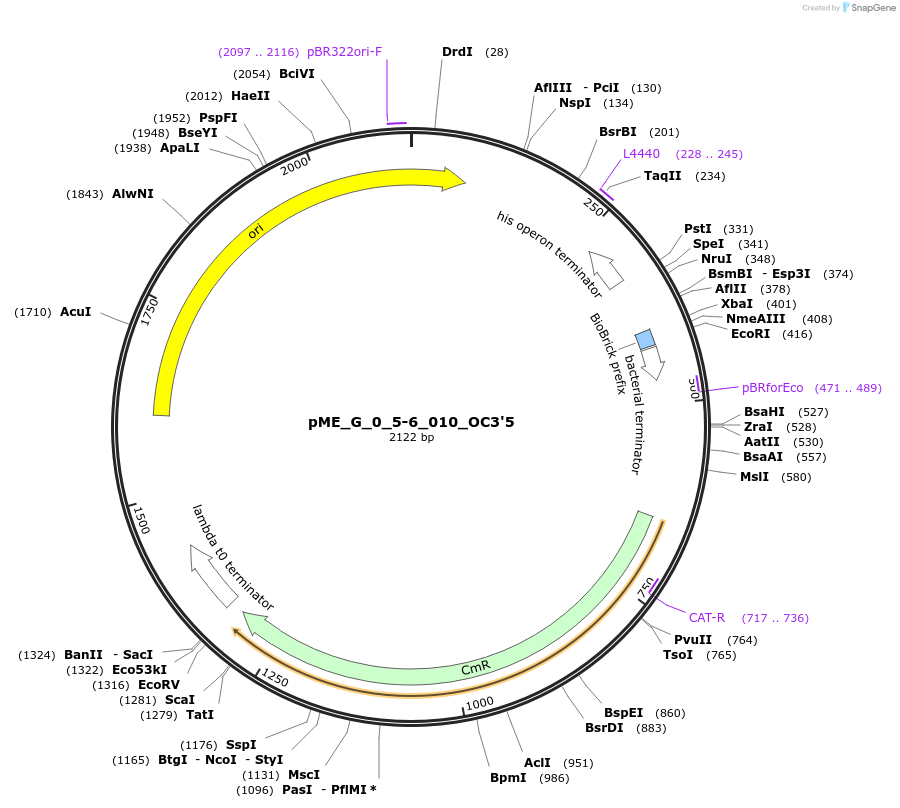
pME_G_0_5-6_010_OC3'5
Plasmid#235966Purpose3' Operon Connector 5 in the M2 position used to construct operonsDepositorInsertOC3'5
UseSynthetic BiologyAvailable SinceMay 5, 2025AvailabilityAcademic Institutions and Nonprofits only