We narrowed to 6,371 results for: KIT;
-
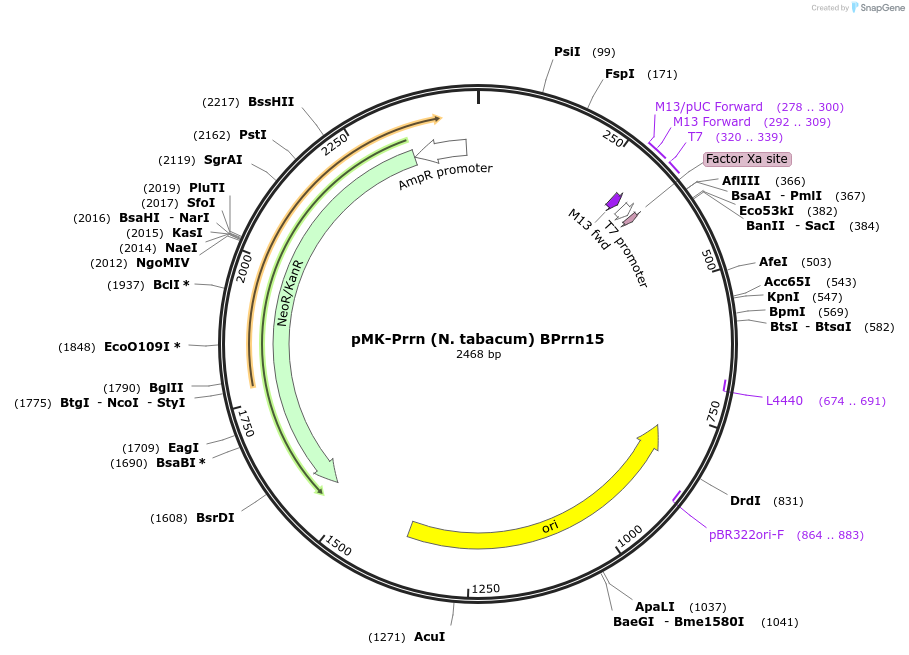
Plasmid#123504PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) BPrrn15
UseSynthetic BiologyAvailable SinceJune 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
pMK-Prrn (N. tabacum) BPrrn14
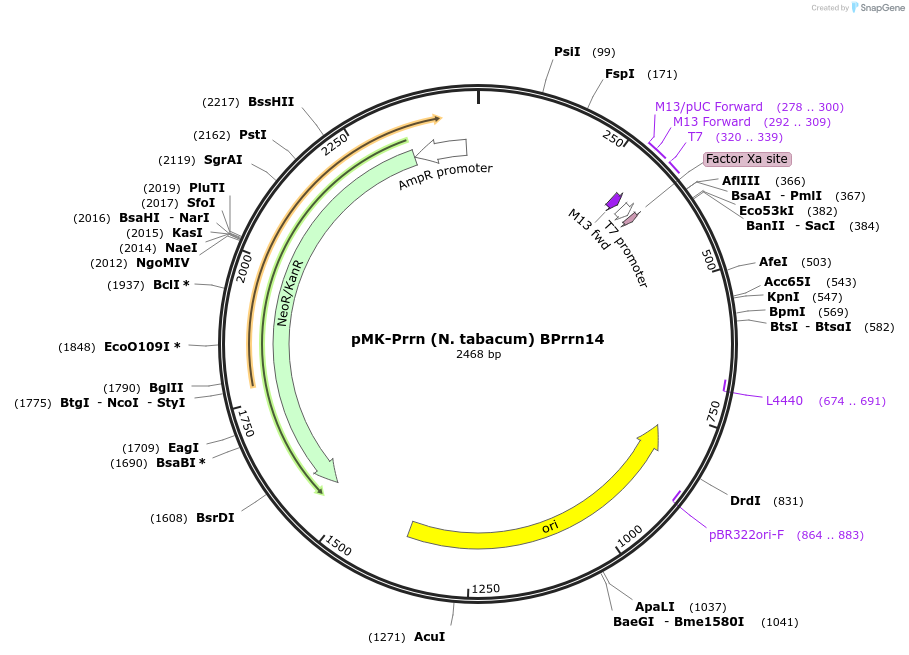
Plasmid#123503PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) BPrrn14
UseSynthetic BiologyAvailable SinceJune 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
pMK-Prrn (N. tabacum) BPrrn13
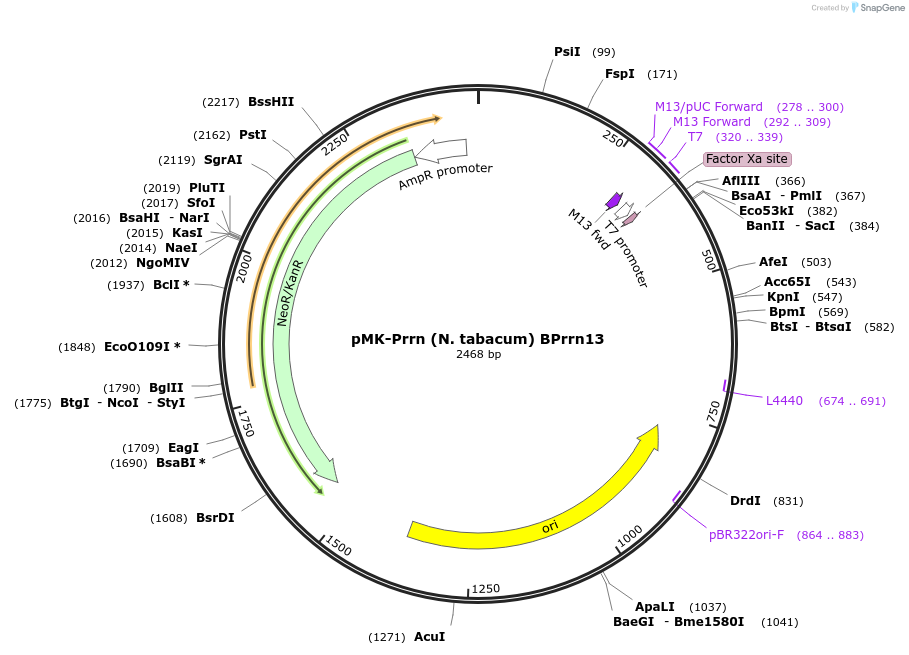
Plasmid#123502PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) BPrrn13
UseSynthetic BiologyAvailable SinceJune 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
pMK-Prrn (N. tabacum) BPrrn12
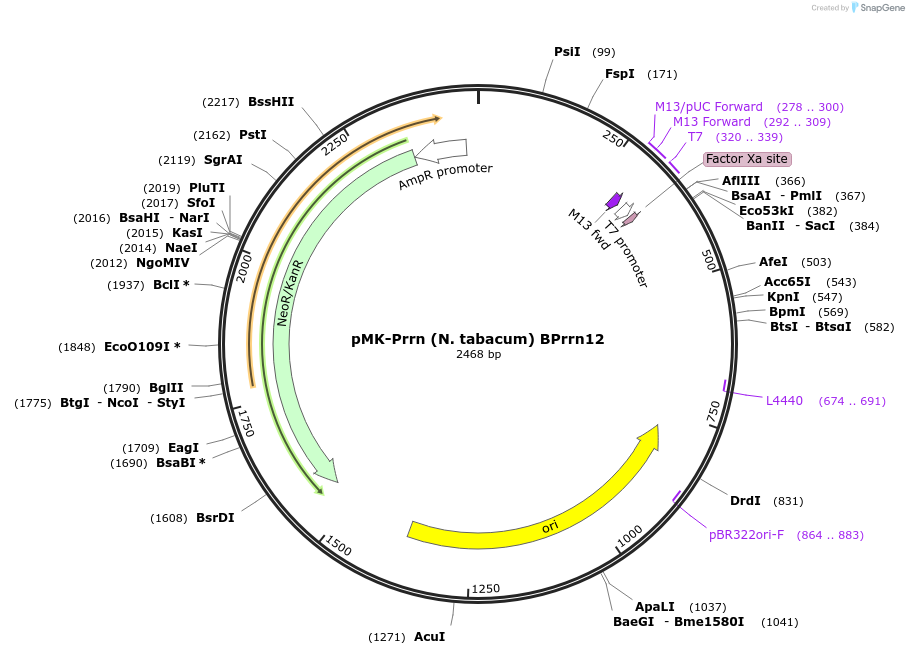
Plasmid#123501PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) BPrrn12
UseSynthetic BiologyAvailable SinceJune 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
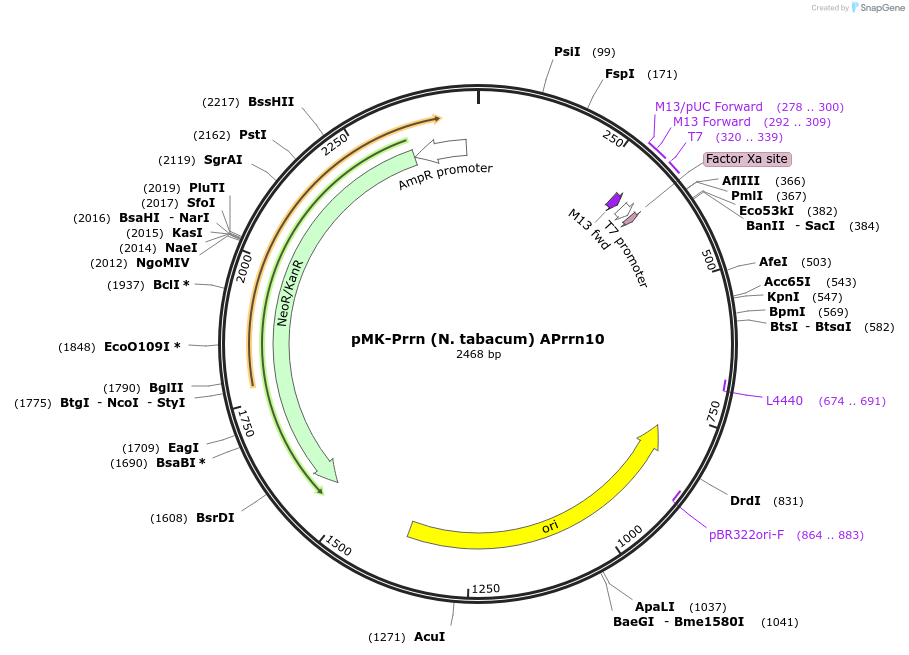
pMK-Prrn (N. tabacum) APrrn10
Plasmid#123500PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) APrrn10
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
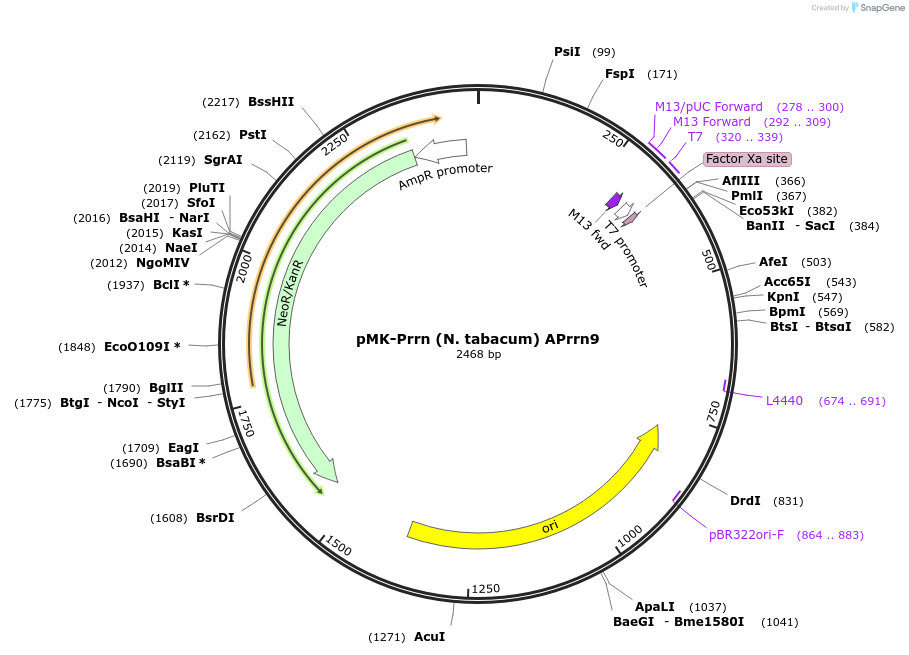
pMK-Prrn (N. tabacum) APrrn9
Plasmid#123499PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) APrrn9
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
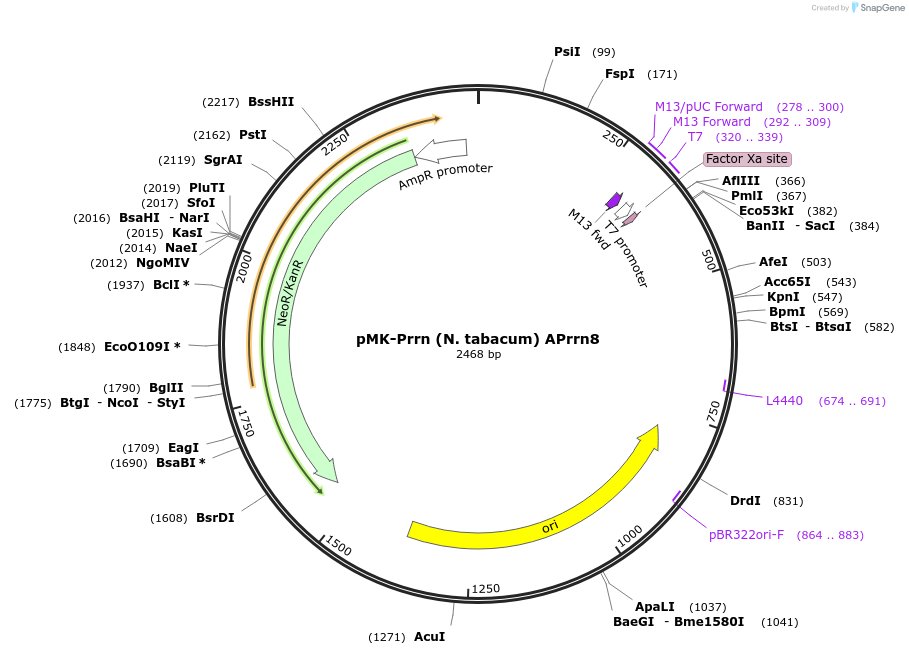
pMK-Prrn (N. tabacum) APrrn8
Plasmid#123498PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) APrrn8
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
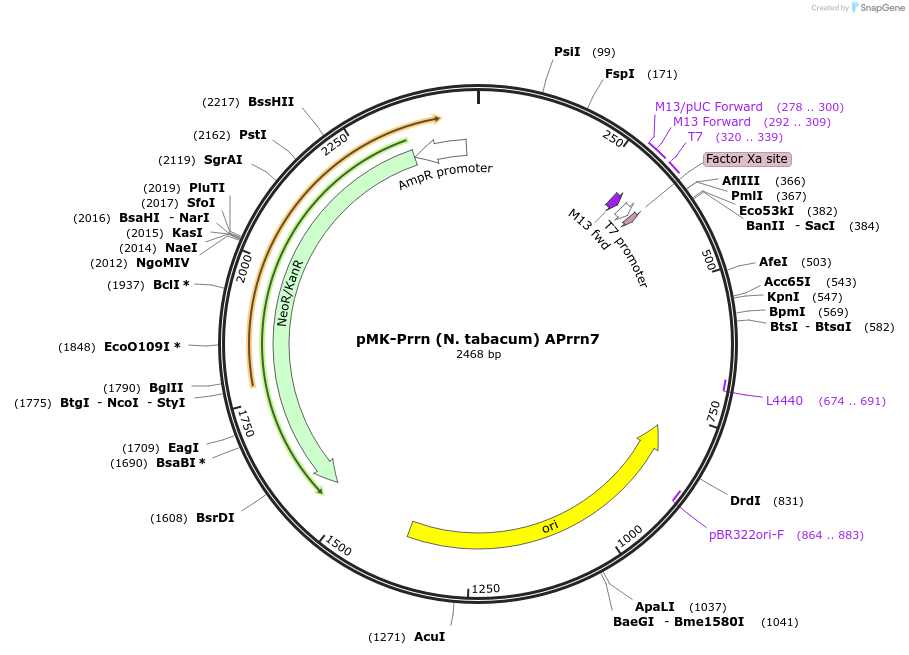
pMK-Prrn (N. tabacum) APrrn7
Plasmid#123497PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) APrrn7
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
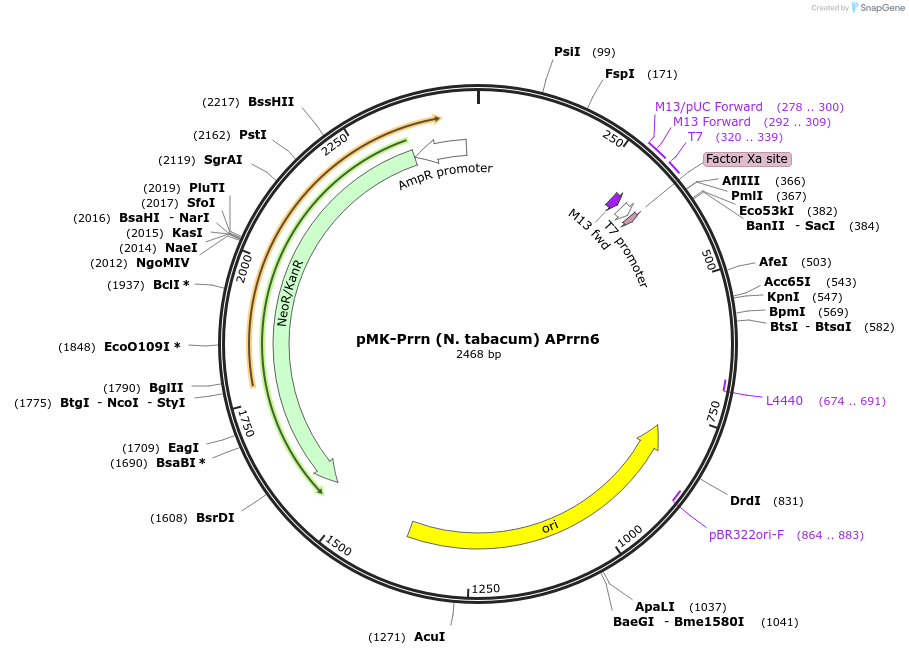
pMK-Prrn (N. tabacum) APrrn6
Plasmid#123496PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) APrrn6
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
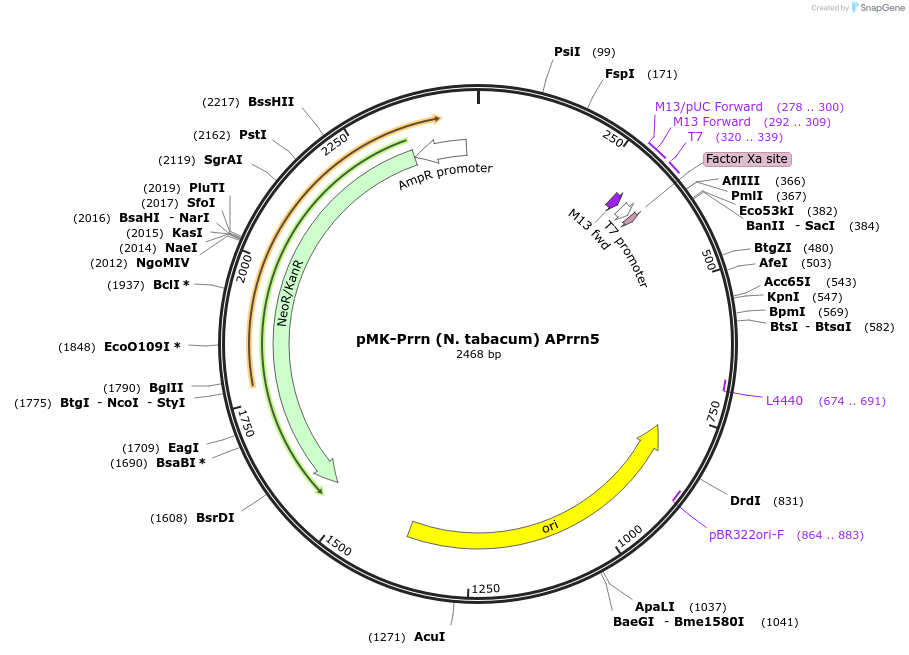
pMK-Prrn (N. tabacum) APrrn5
Plasmid#123495PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) APrrn5
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
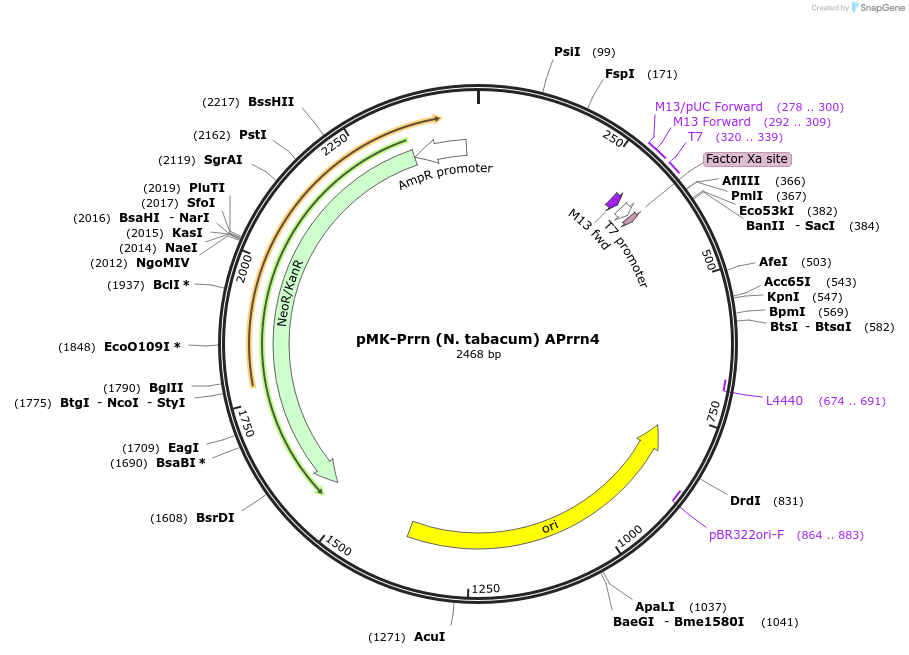
pMK-Prrn (N. tabacum) APrrn4
Plasmid#123494PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) APrrn4
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
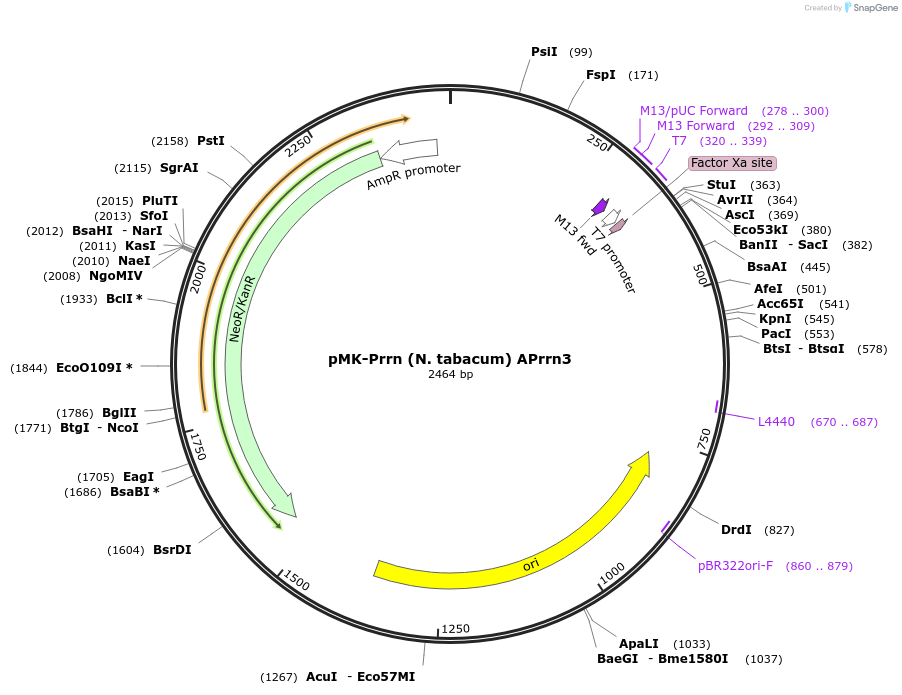
pMK-Prrn (N. tabacum) APrrn3
Plasmid#123493PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) APrrn3
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
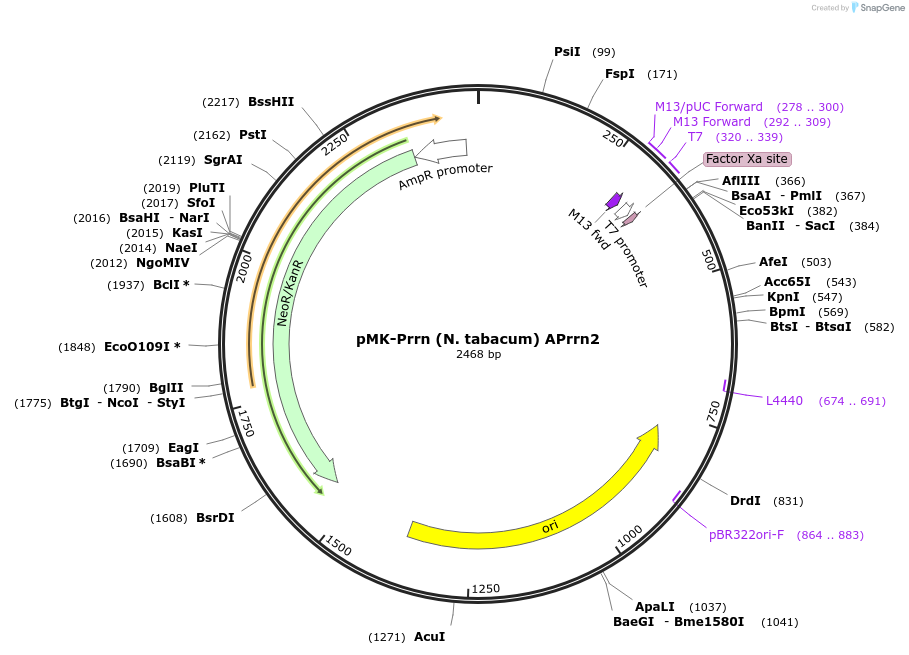
pMK-Prrn (N. tabacum) APrrn2
Plasmid#123492PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) APrrn2
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
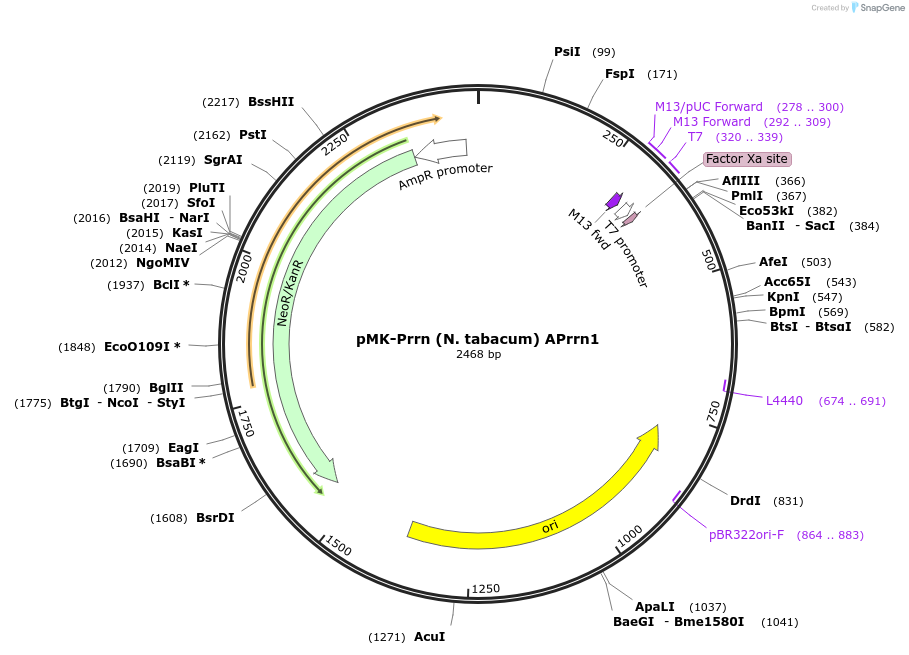
pMK-Prrn (N. tabacum) APrrn1
Plasmid#123491PurposeMoChlo kitDepositorInsertPrrn (N. tabacum) APrrn1
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
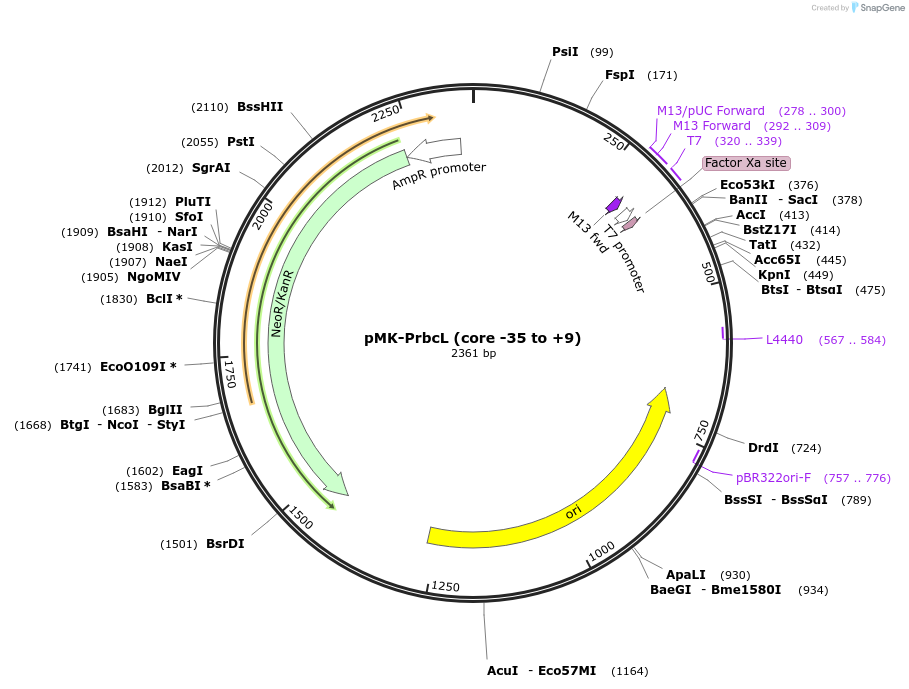
pMK-PrbcL (core -35 to +9)
Plasmid#123486PurposeMoChlo kitDepositorInsertPrbcL (core -35 to +9)
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
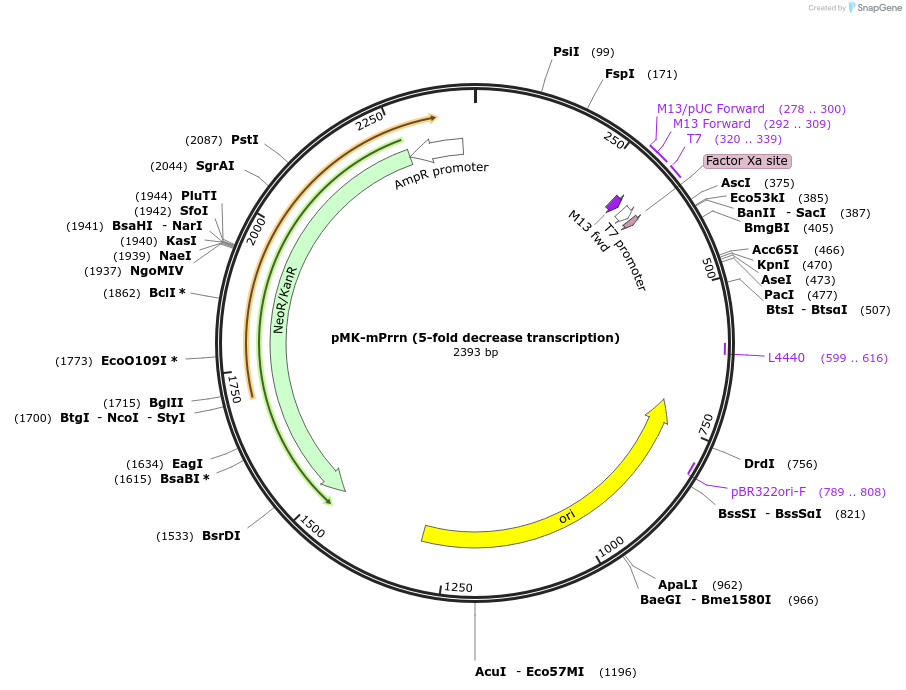
pMK-mPrrn (5-fold decrease transcription)
Plasmid#123478PurposeMoChlo kitDepositorInsertmPrrn (5-fold decrease transcription)
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
pMK-PatpB (PEP functional NEP disabled)
Plasmid#123469PurposeMoChlo kitDepositorInsertPatpB (PEP functional NEP disabled)
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
pMK-PatpB (PEP and NEP functional)
Plasmid#123468PurposeMoChlo kitDepositorInsertPatpB (PEP and NEP functional)
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
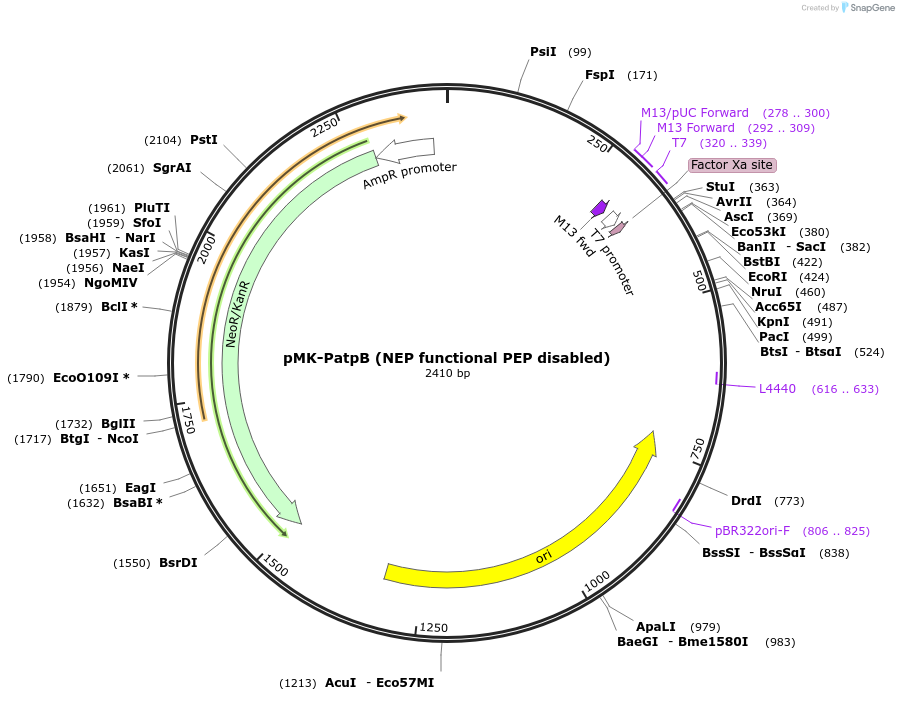
pMK-PatpB (NEP functional PEP disabled)
Plasmid#123466PurposeMoChlo kitDepositorInsertPatpB (NEP functional PEP disabled)
UseSynthetic BiologyAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
pDONR223-LOC652799
Plasmid#23394DepositorAvailable SinceJuly 30, 2010AvailabilityAcademic Institutions and Nonprofits only