We narrowed to 11,363 results for: ENA;
-
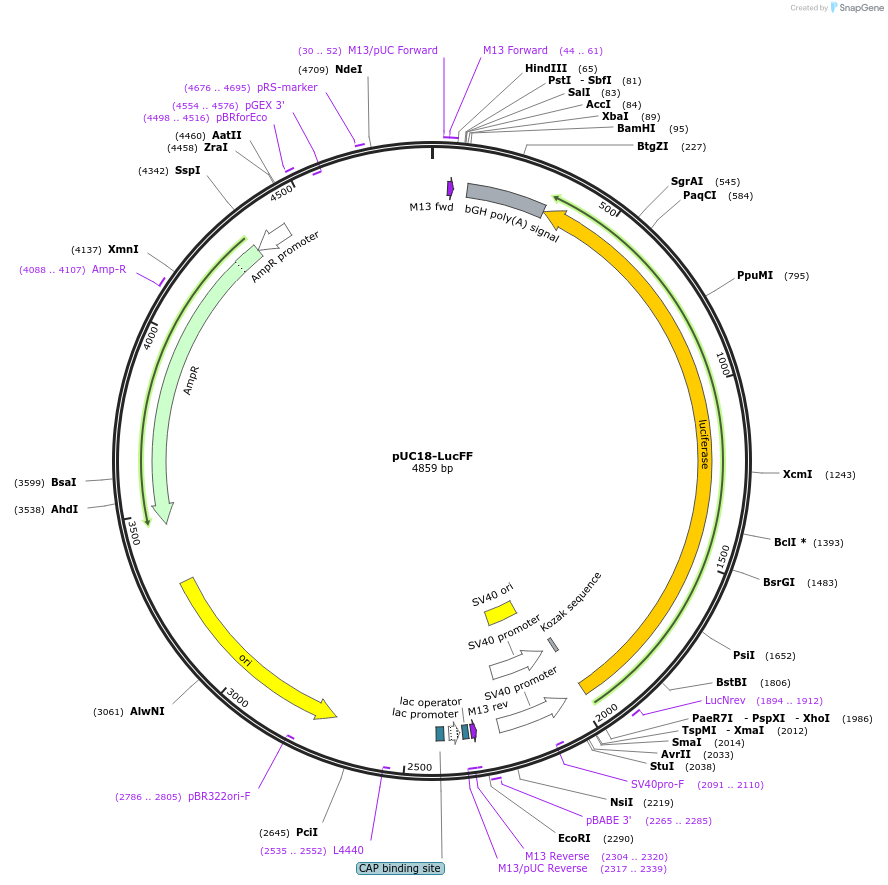
Plasmid#220496PurposeTo generate substrates to monitor Ku overloading and its impact on transcription at the DNA ends vicinity.DepositorInsertFirefly Luciferase
ExpressionMammalianPromoterSV40Available SinceJuly 3, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
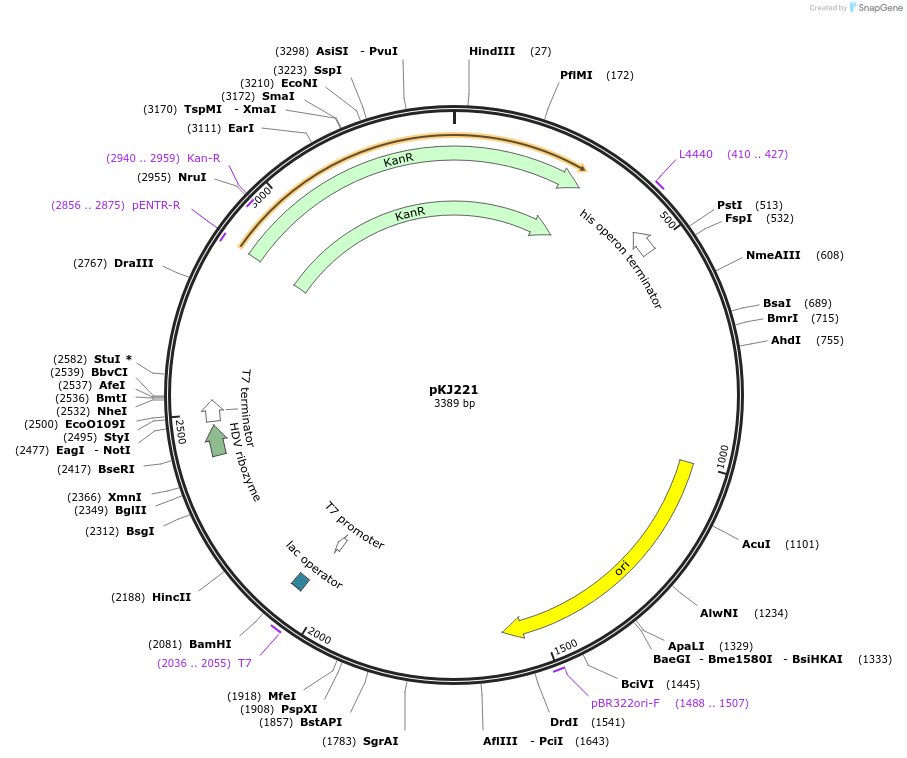
pKJ221
Plasmid#212319PurposeExpress MmFnuc guide with pureexpressDepositorInsertMercenaria guide
ExpressionBacterialAvailable SinceApril 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
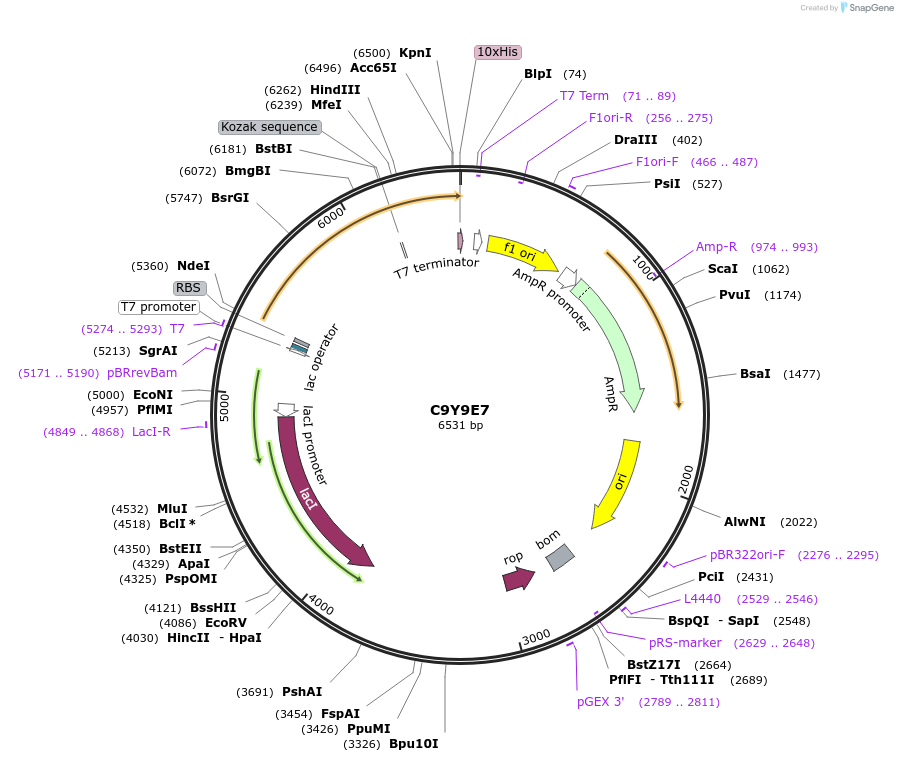
C9Y9E7
Plasmid#163230PurposeInducible expression of UniProt identifier C9Y9E7DepositorInsertC9Y9E7
Tags10xHis and T7ExpressionBacterialPromoterT7Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
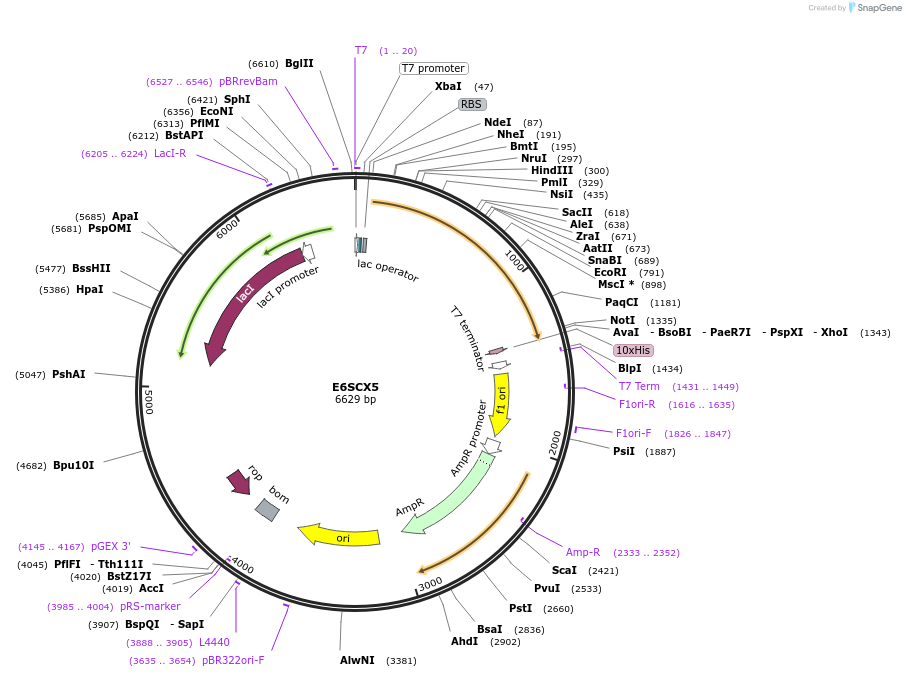
E6SCX5
Plasmid#163275PurposeInducible expression of UniProt identifier E6SCX5DepositorInsertE6SCX5
Tags10xHis and T7ExpressionBacterialPromoterT7Available SinceFeb. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
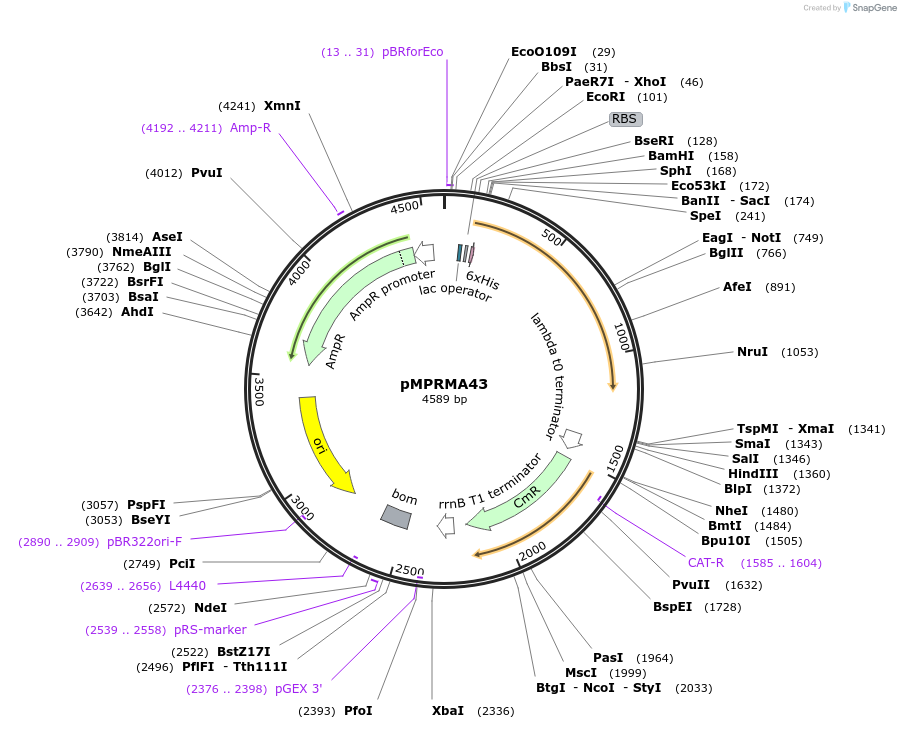
pMPRMA43
Plasmid#172944PurposepQE30 encoding Wzz mutant i191DepositorInsertShigella flexneri wzz i191 mutant gene
Tags6xHisExpressionBacterialAvailable SinceSept. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
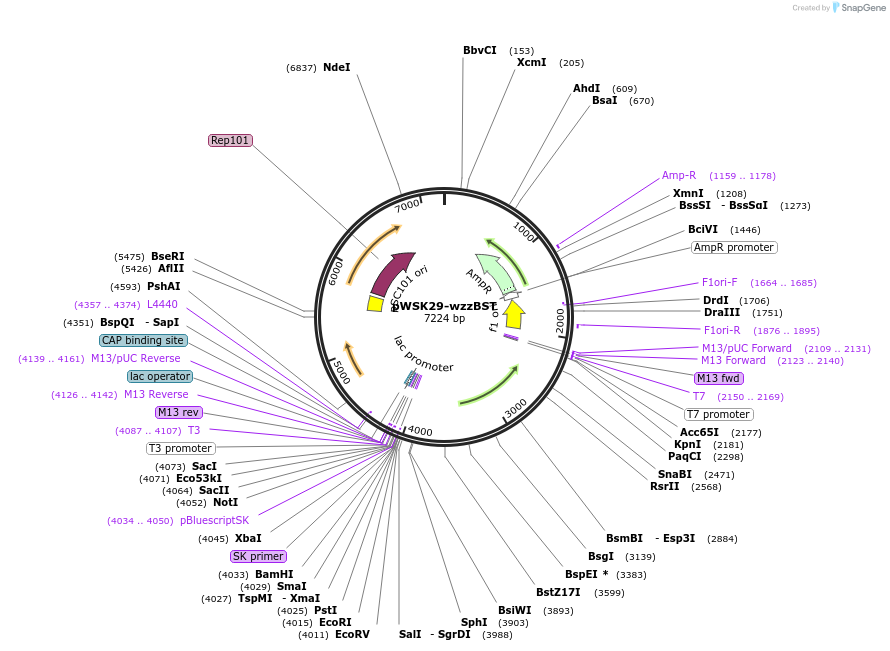
pWSK29-wzzBST
Plasmid#172968PurposepWSK29 with Salmonella typhimurium wzzBST geneDepositorInsertwzzB gene from Salmonella enterica serovar Typhimurium
ExpressionBacterialMutationK198E- please see depositor commentsAvailable SinceAug. 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
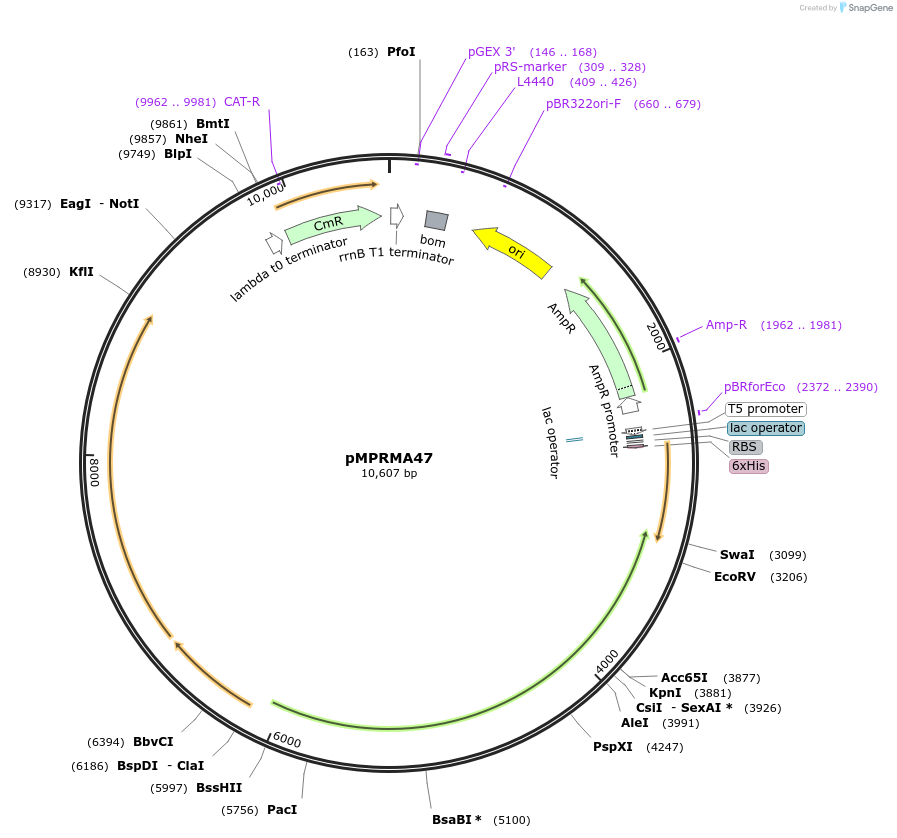
pMPRMA47
Plasmid#172947PurposepQE30 encoding Wzz mutant i255DepositorInsertShigella flexneri wzz i255 mutant gene
Tags6xHisExpressionBacterialAvailable SinceAug. 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
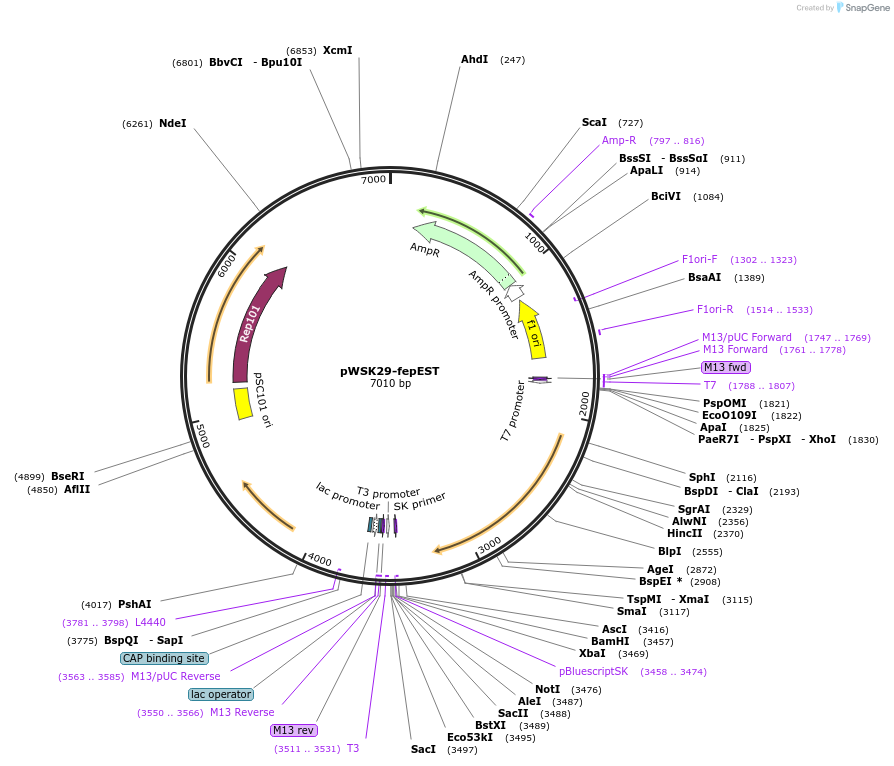
pWSK29-fepEST
Plasmid#172969PurposepWSK29 with Salmonella typhimurium fepEST geneDepositorInsertfepE gene from Salmonella enterica serovar Typhimurium
ExpressionBacterialMutationG160S- please see depositor commentsAvailable SinceAug. 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
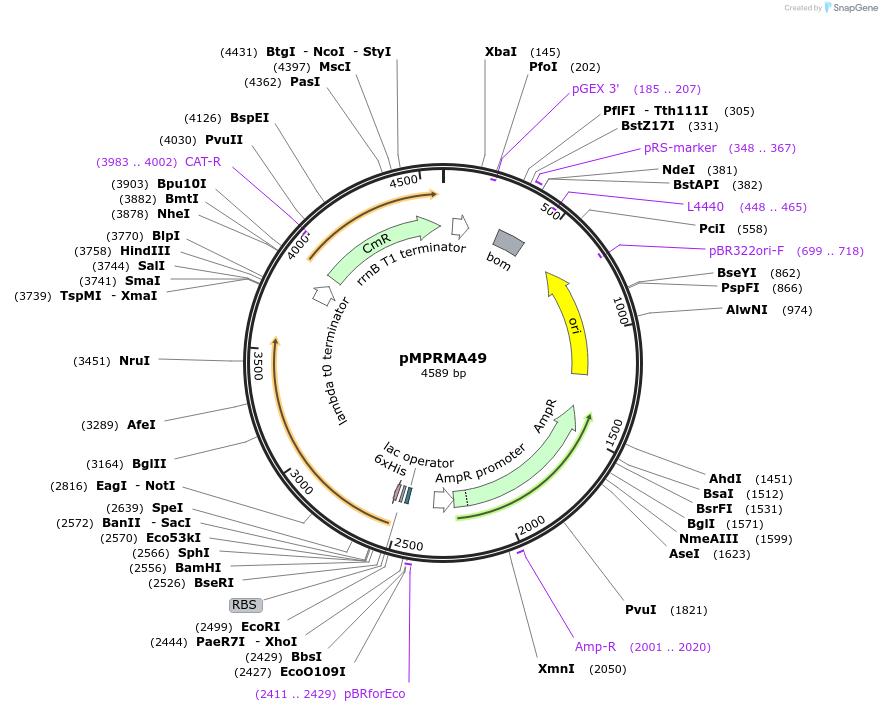
pMPRMA49
Plasmid#172948PurposepQE30 encoding Wzz mutant i81DepositorInsertShigella flexneri wzz i81 mutant gene
Tags6xHisExpressionBacterialAvailable SinceAug. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
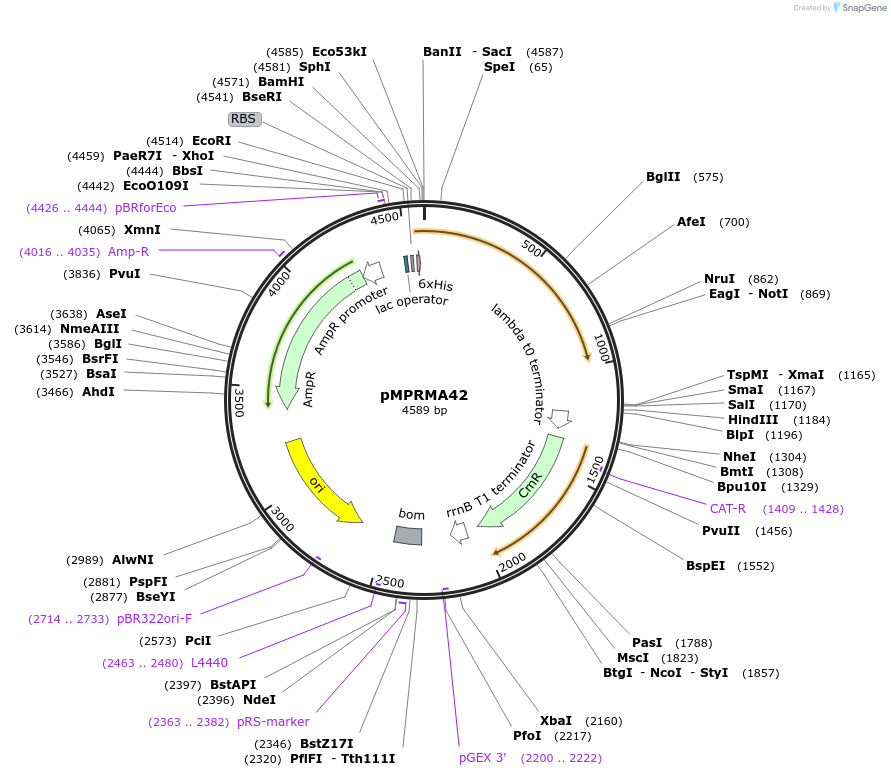
pMPRMA42
Plasmid#172943PurposepQE30 encoding Wzz mutant i290DepositorInsertShigella flexneri wzz i290 mutant gene
Tags6xHisExpressionBacterialAvailable SinceAug. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
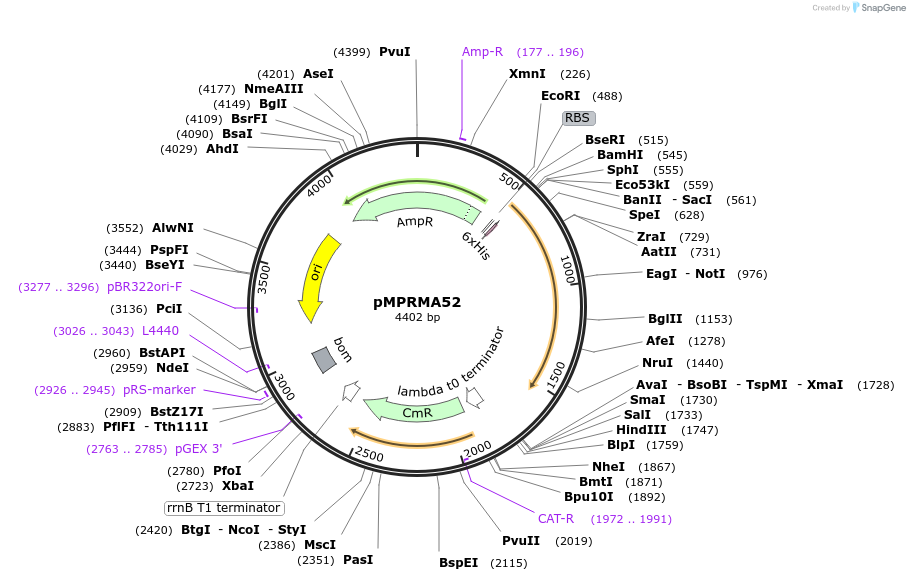
pMPRMA52
Plasmid#172938PurposepQE30 encoding Wzz mutant i138DepositorInsertShigella flexneri wzz i138 mutant gene
Tags6xHisExpressionBacterialAvailable SinceAug. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
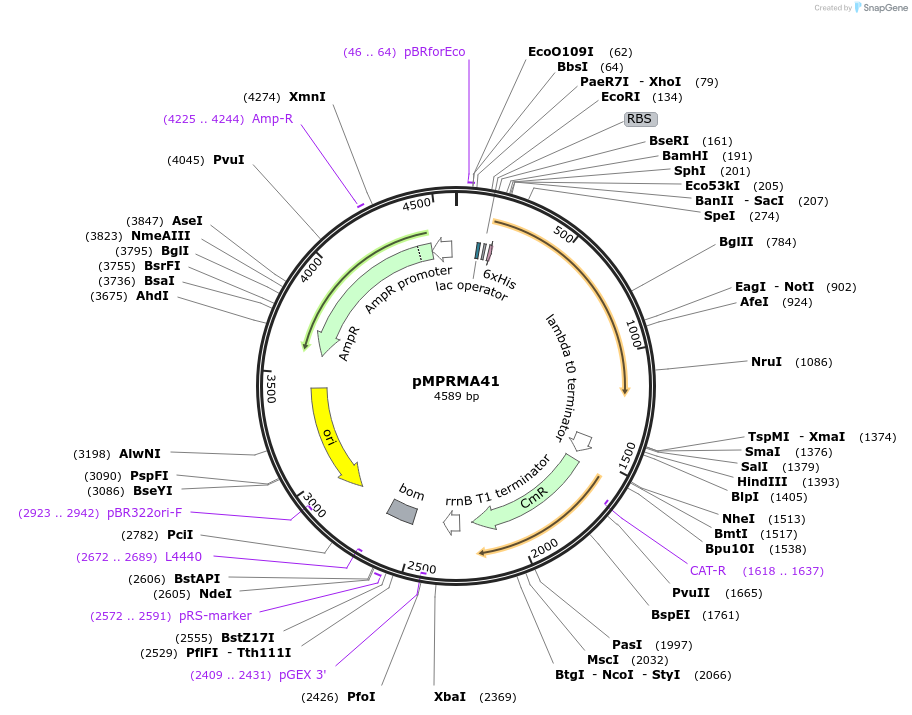
pMPRMA41
Plasmid#172935PurposepQE30 encoding Wzz mutant i231DepositorInsertShigella flexneri wzz i231 mutant gene
Tags6xHisExpressionBacterialAvailable SinceAug. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
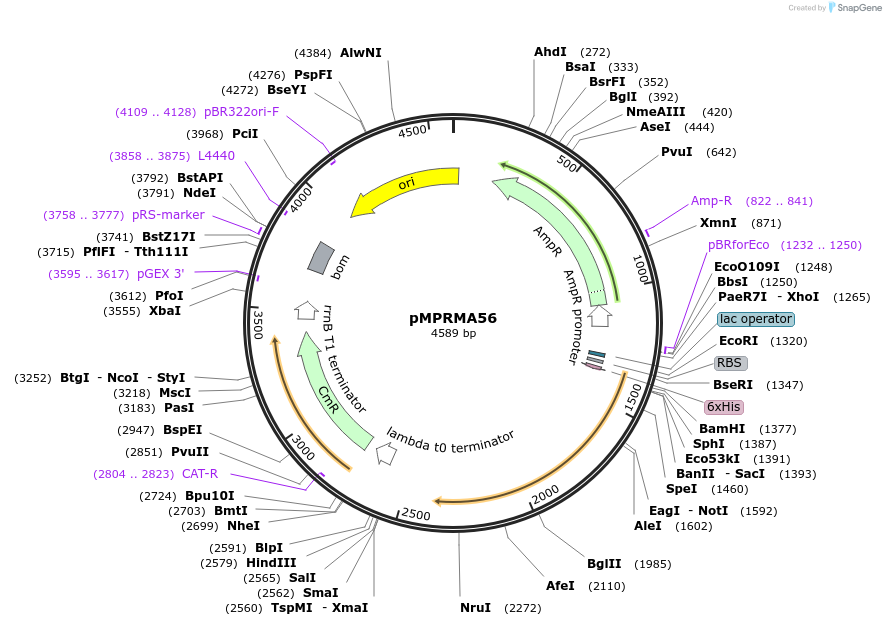
pMPRMA56
Plasmid#172951PurposepQE30 encoding Wzz mutant i66DepositorInsertShigella flexneri wzz i66 mutant gene
Tags6xHisExpressionBacterialAvailable SinceAug. 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
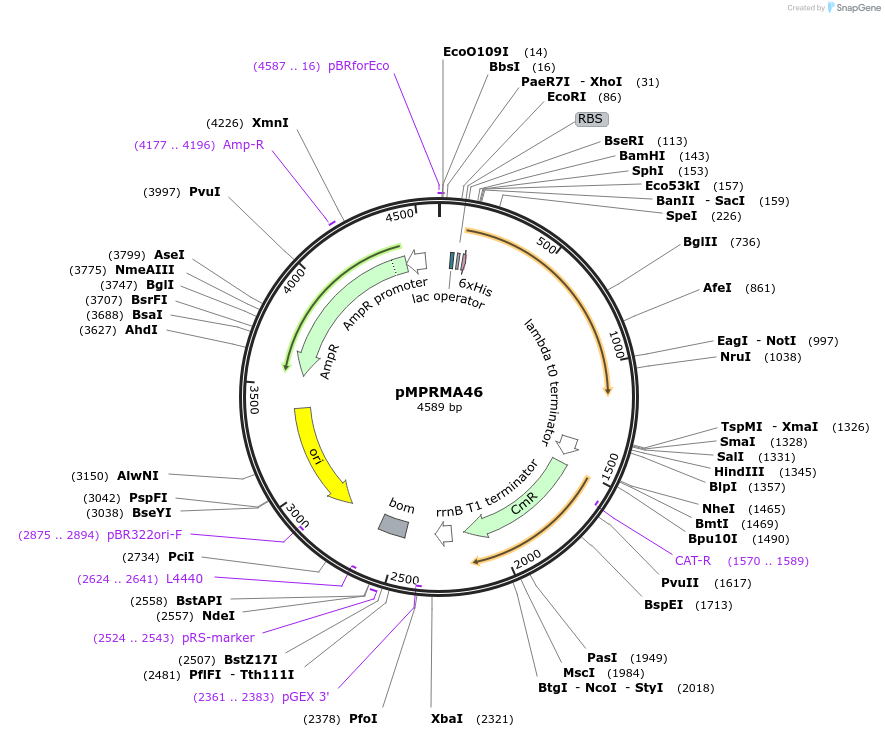
pMPRMA46
Plasmid#172946PurposepQE30 encoding Wzz mutant i279DepositorInsertShigella flexneri wzz i279 mutant gene
Tags6xHisExpressionBacterialAvailable SinceAug. 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
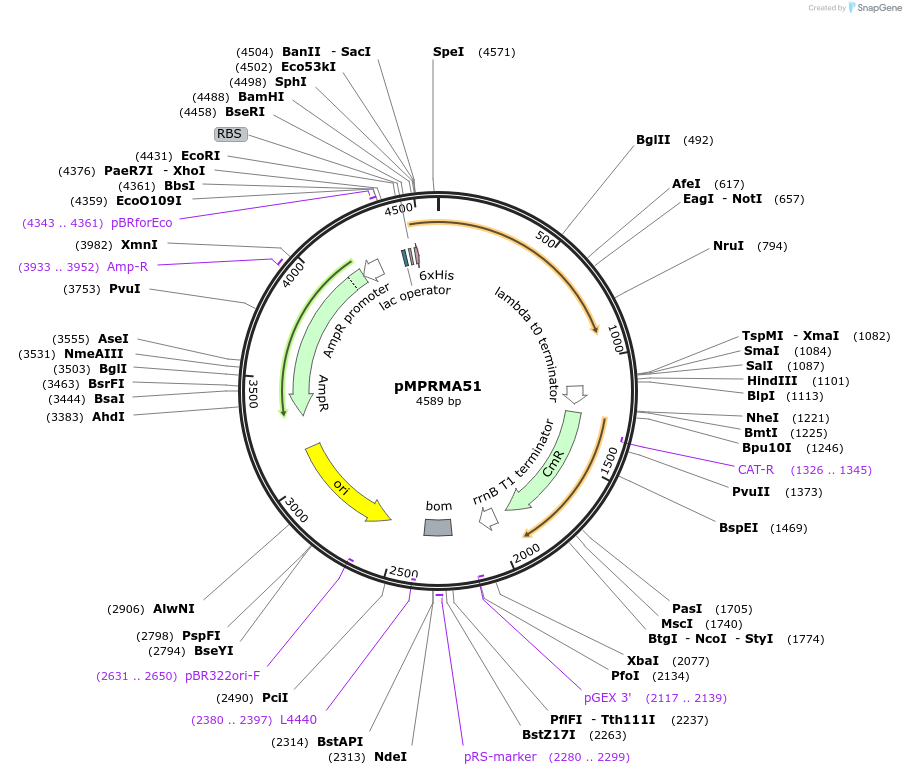
pMPRMA51
Plasmid#172949PurposepQE30 encoding Wzz mutant i247DepositorInsertShigella flexneri wzz i247 mutant gene
Tags6xHisExpressionBacterialAvailable SinceAug. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
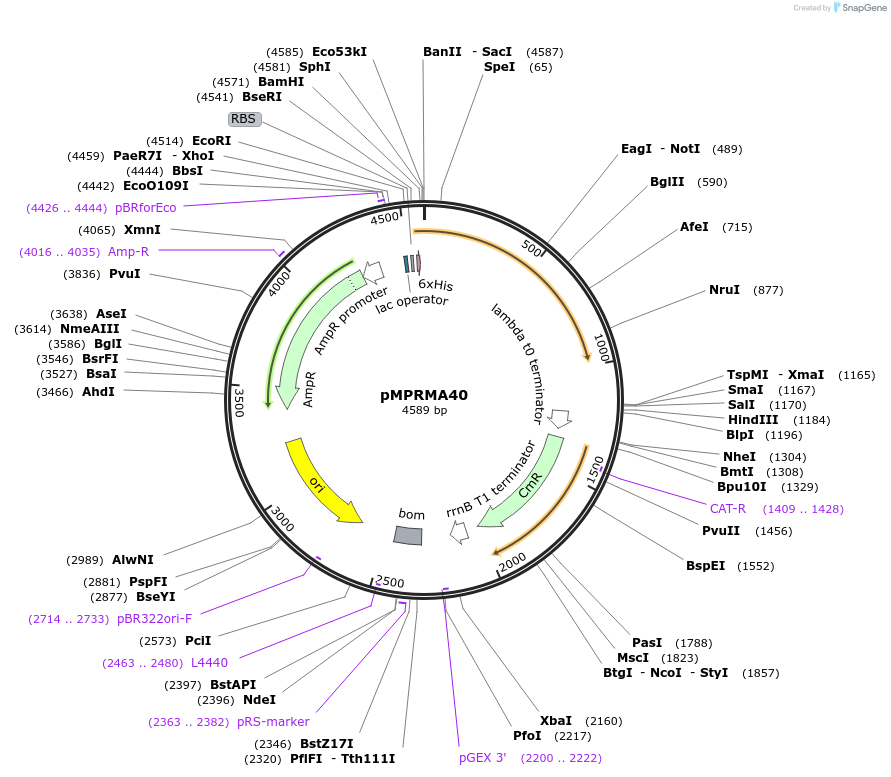
pMPRMA40
Plasmid#172942PurposepQE30 encoding Wzz mutant i163DepositorInsertShigella flexneri wzz i163 mutant gene
Tags6xHisExpressionBacterialAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
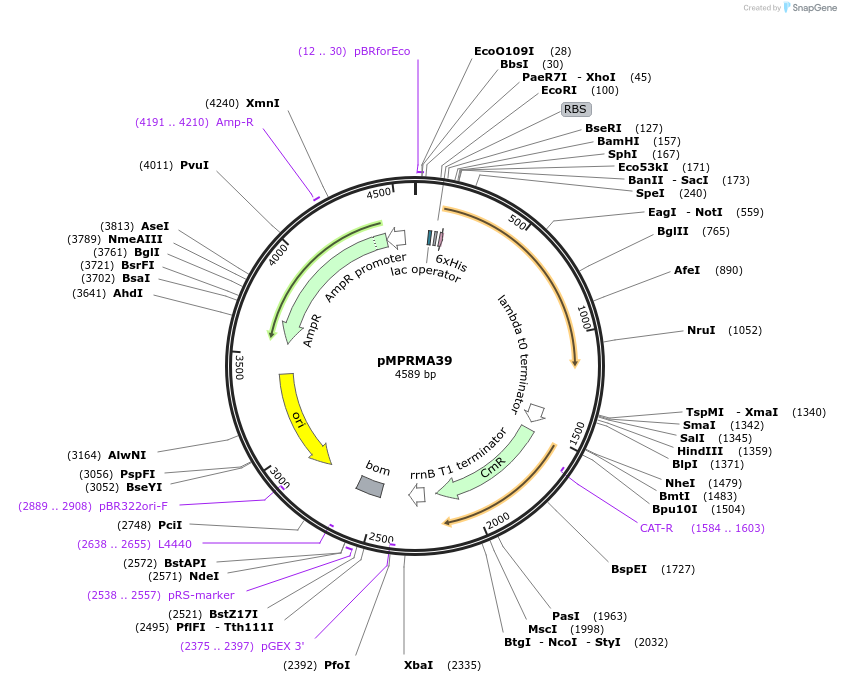
pMPRMA39
Plasmid#172940PurposepQE30 encoding Wzz mutant i128DepositorInsertShigella flexneri wzz i128 mutant gene
Tags6xHisExpressionBacterialAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
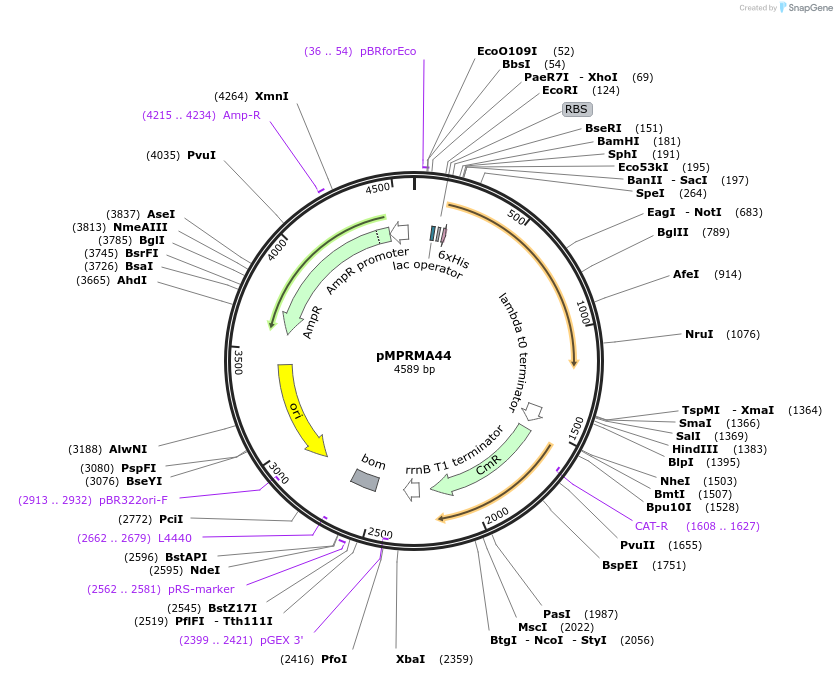
pMPRMA44
Plasmid#172945PurposepQE30 encoding Wzz mutant i161DepositorInsertShigella flexneri wzz i161 mutant gene
Tags6xHisExpressionBacterialAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
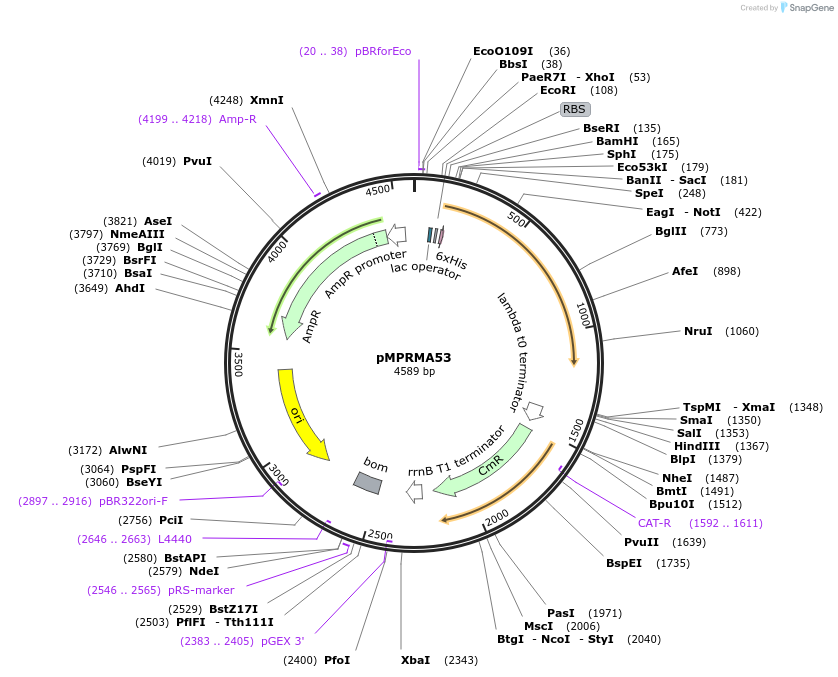
pMPRMA53
Plasmid#172939PurposepQE30 encoding Wzz mutant i80DepositorInsertShigella flexneri wzz i80 mutant gene
Tags6xHisExpressionBacterialAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
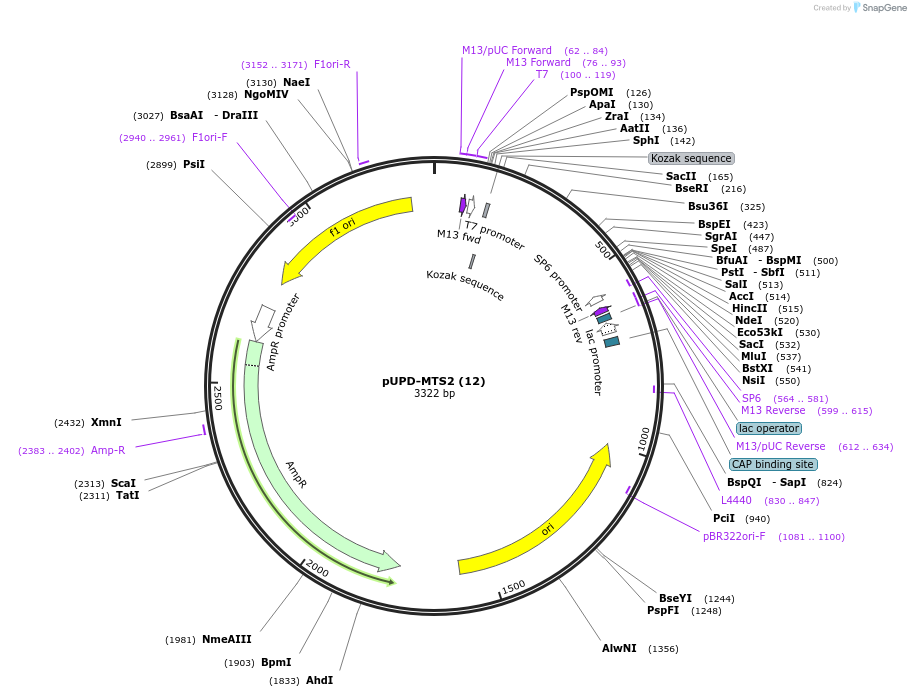
pUPD-MTS2 (12)
Plasmid#101698PurposeMTS2 in domestication vectorDepositorInsertCCAT-[MTS from β subunit of Nicotiana plumbaginifolia F1 ATPase]-AATG
UseYeast expressionAvailable SinceJuly 24, 2018AvailabilityAcademic Institutions and Nonprofits only