We narrowed to 4,849 results for: Kl;
-
Plasmid#243697PurposeA piggyBac vector containing the endogenous NDUFB9 genomic locus, with its native 5' UTR replaced by the Actb 5' UTR, modified with silent mutations to allow for PCR-based analysisDepositorInsertNDUFB9 genomic locus, with its 5' UTR replaced by the ACTB 5' UTR (NDUFB9 Human)
ExpressionMammalianAvailable SinceNov. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
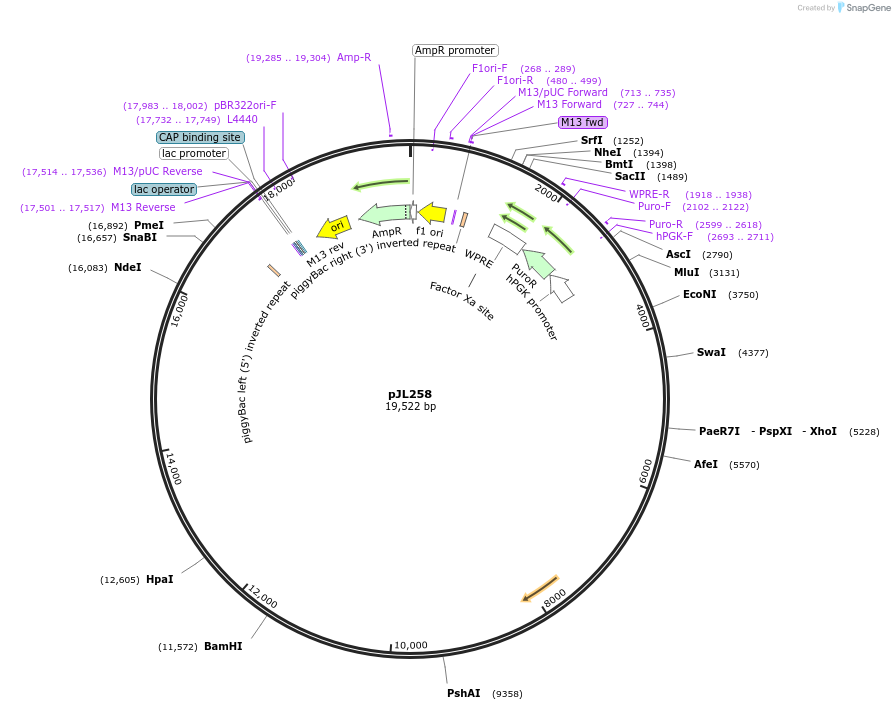
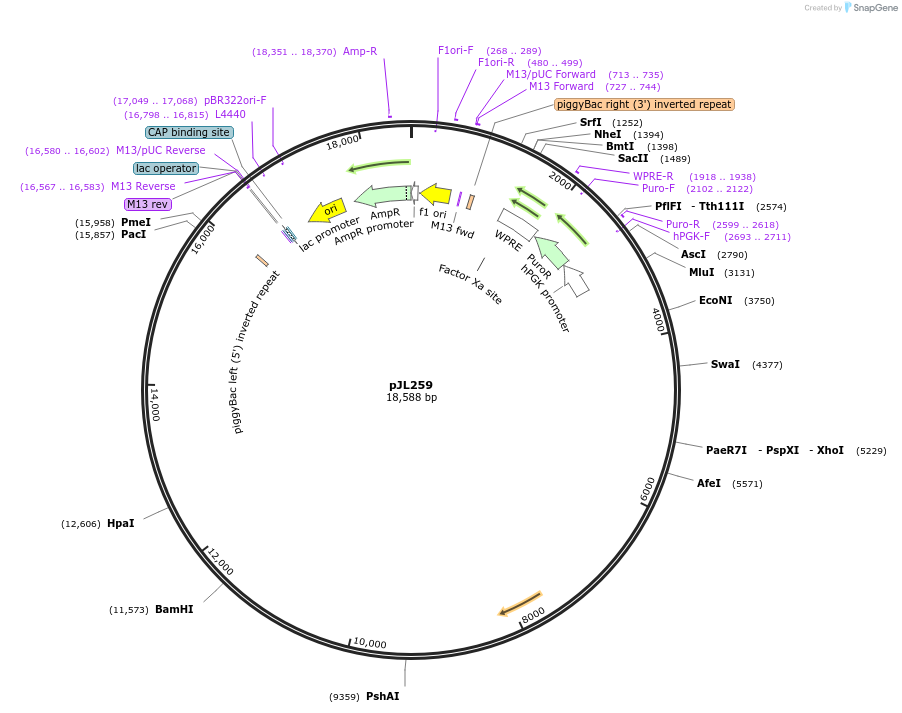
pJL259
Plasmid#243698PurposeA piggyBac vector containing the endogenous NDUFB9 genomic locus, with its native 3' UTR replaced by the Actb 3' UTR, modified with silent mutations to allow for PCR-based analysisDepositorInsertNDUFB9 genomic locus, with its 3' UTR replaced by the ACTB 3' UTR (NDUFB9 Human)
ExpressionMammalianAvailable SinceNov. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
-
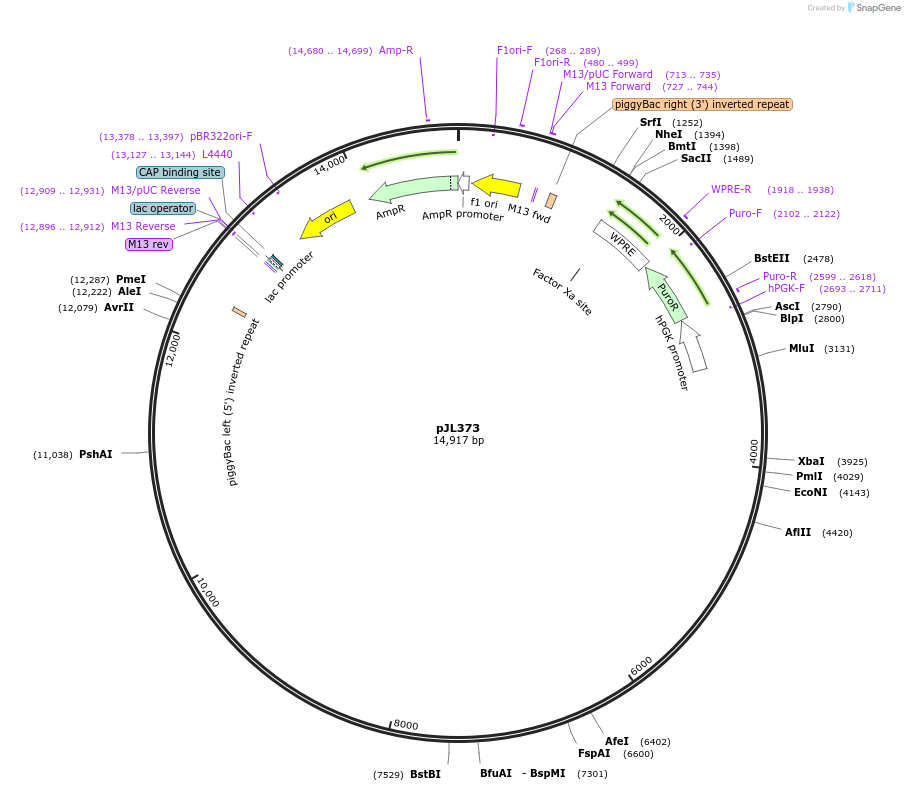
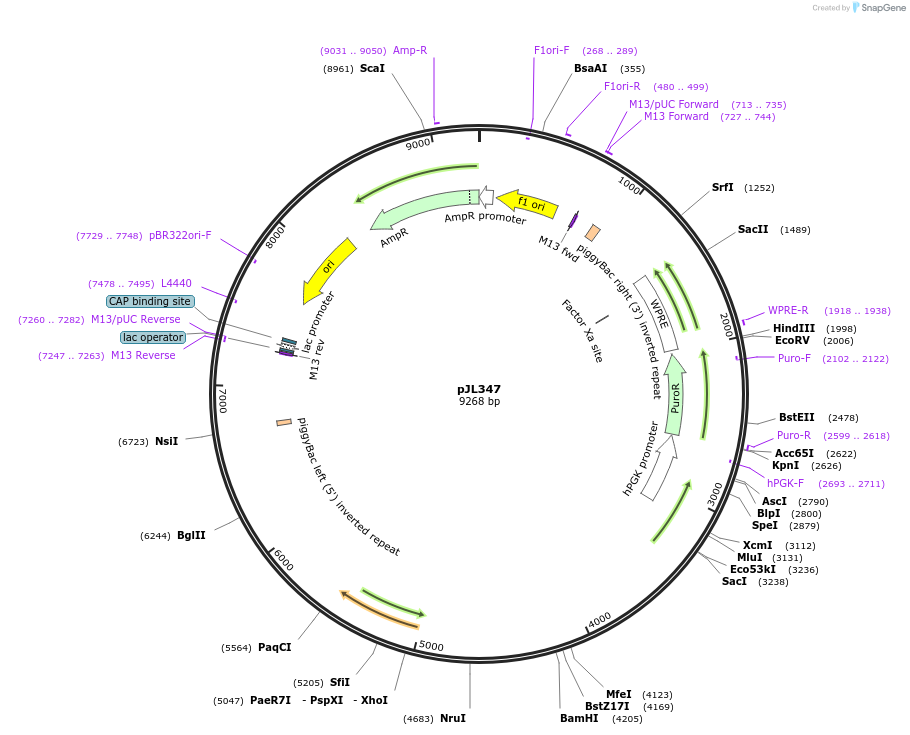
pJL347
Plasmid#243707PurposeA piggyBac vector containing the COX5B coding sequences, modified with silent mutations to allow for PCR-based analysis, along with endogenous promoter and UTRs.DepositorAvailable SinceNov. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
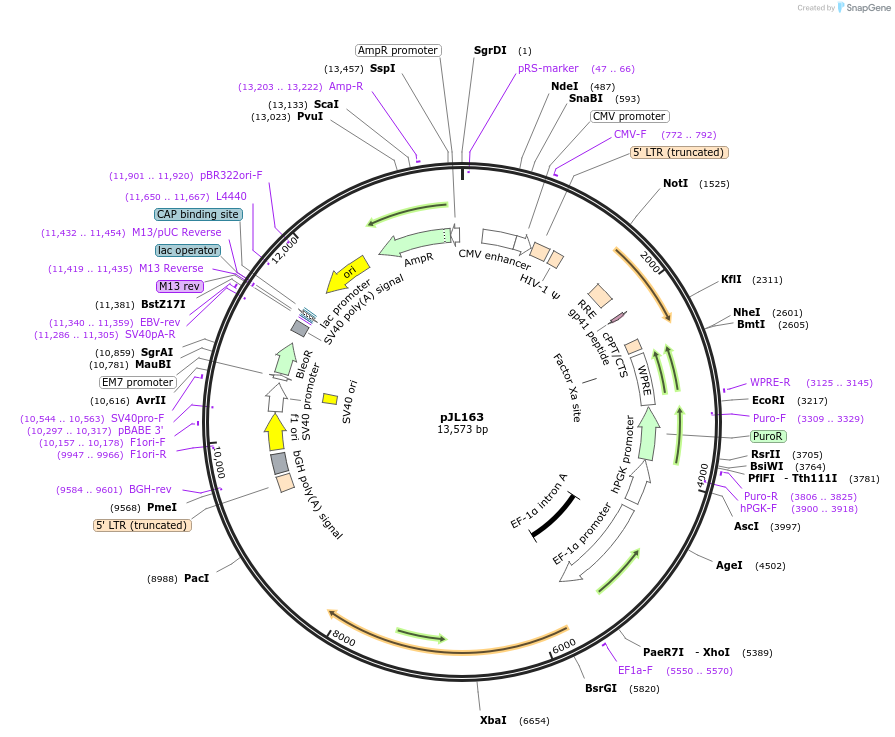
pJL163
Plasmid#243686PurposeStably expresses ALDH18A1 CDS with its endogenous 5'UTR and 3'UTRDepositorAvailable SinceNov. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
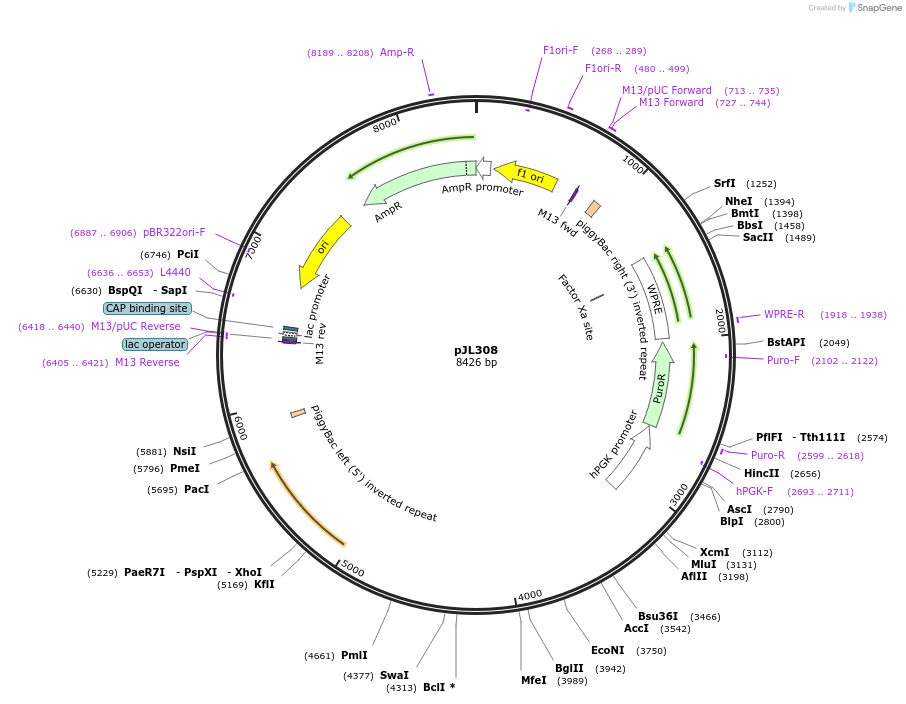
pJL308
Plasmid#243704PurposeA piggyBac vector containing the NDUFB9 coding sequence under the control of its endogenous promoter and 5' UTR, followed by the ACTB 3' UTRDepositorInsertNDUFB9 coding sequence with endogenous 5' UTR, followed by the ACTB 3' UTR (NDUFB9 Human)
ExpressionMammalianPromoterendogenous promoterAvailable SinceOct. 15, 2025AvailabilityAcademic Institutions and Nonprofits only -
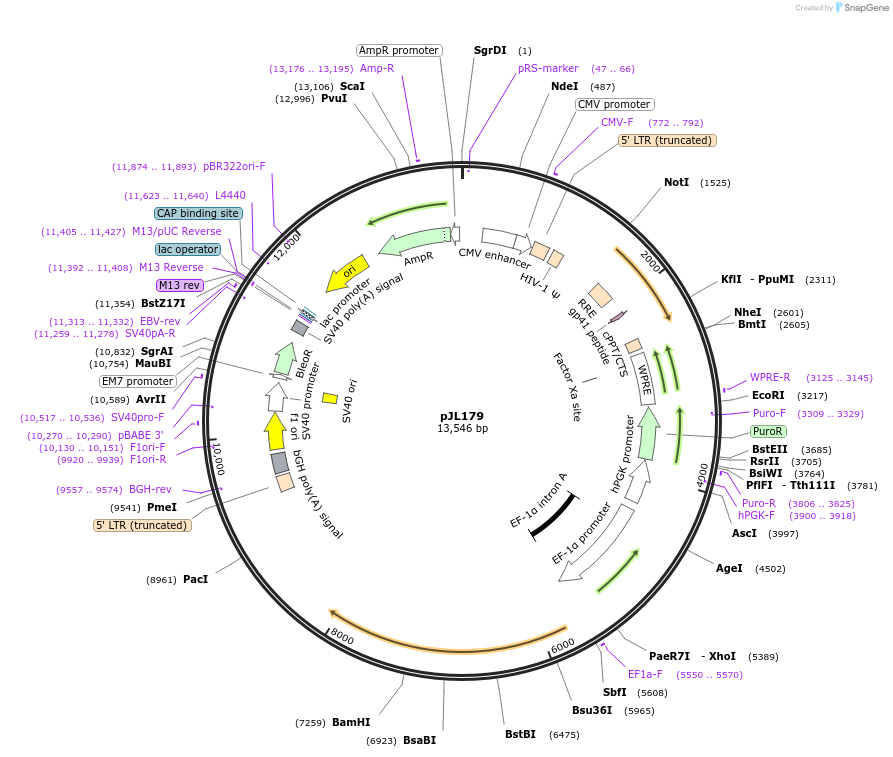
pJL179
Plasmid#243688PurposeStably expresses SUPV3L1 CDS, flanked with endogenous 5' and 3'UTRs of ALDH18A1DepositorAvailable SinceOct. 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
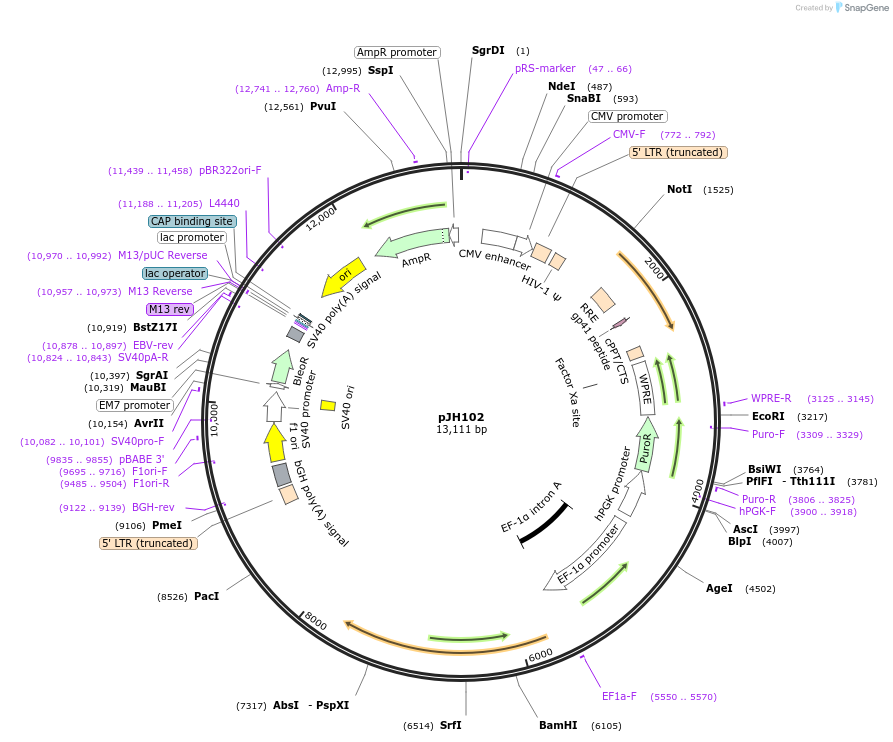
pJH102
Plasmid#243725PurposeExpresses a chimeric form of TBRG4, where the region between residue 110 and 351 is replaced by a 250-residue XTEN linkerDepositorInsertTBRG4(1-110aa)-XTEN250-TBRG4(351aa-stop) (TBRG4 Human)
UseLentiviralMutationThe region between residue 110 and 351 of TBRG4 i…Available SinceOct. 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
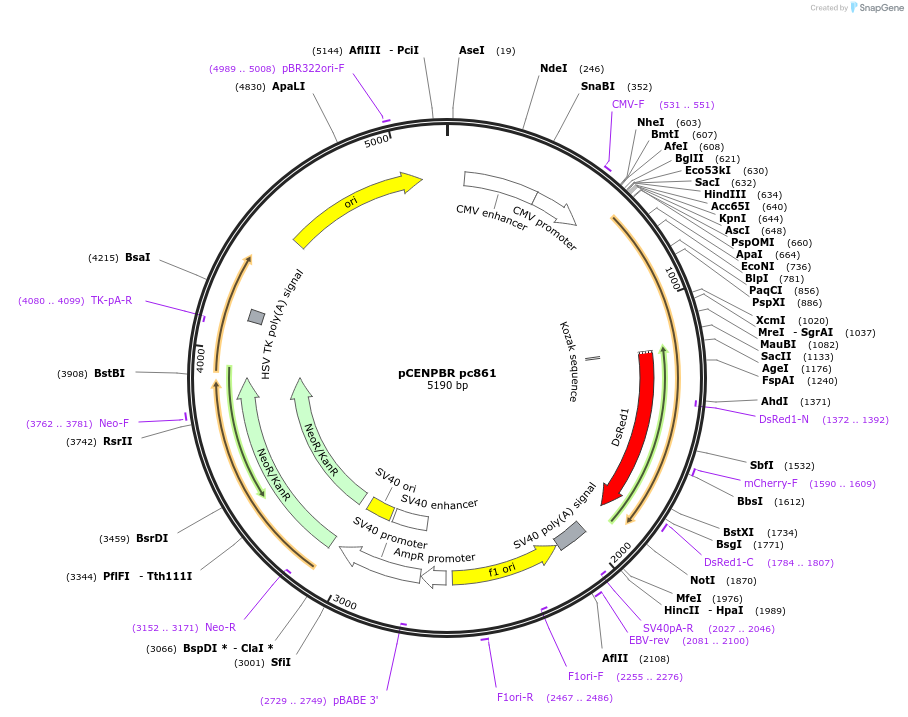
pCENPBR pc861
Plasmid#167569PurposeHuman CENP-B (aa 1-169 containing DNA-binding domain) cloned into pDsRed1-N1 vector (Clontech).DepositorInsertHomo sapiens centromere protein B (CENPB Human)
TagsDs-RedExpressionMammalianPromoterCMVAvailable SinceJune 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
pGreenII 0229/ProRUBY:GOXL6-Citrine:NOSt
Plasmid#175568PurposepSOUP helper plasmid needed for Agrobacterium-mediated plant transformation - Expression of GOXL genes tagged with Citrine under RUBY promoter for transgene complementation of ruby-6DepositorInsertGOXL6
TagsCitrineExpressionPlantPromoterRUBYAvailable SinceJan. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
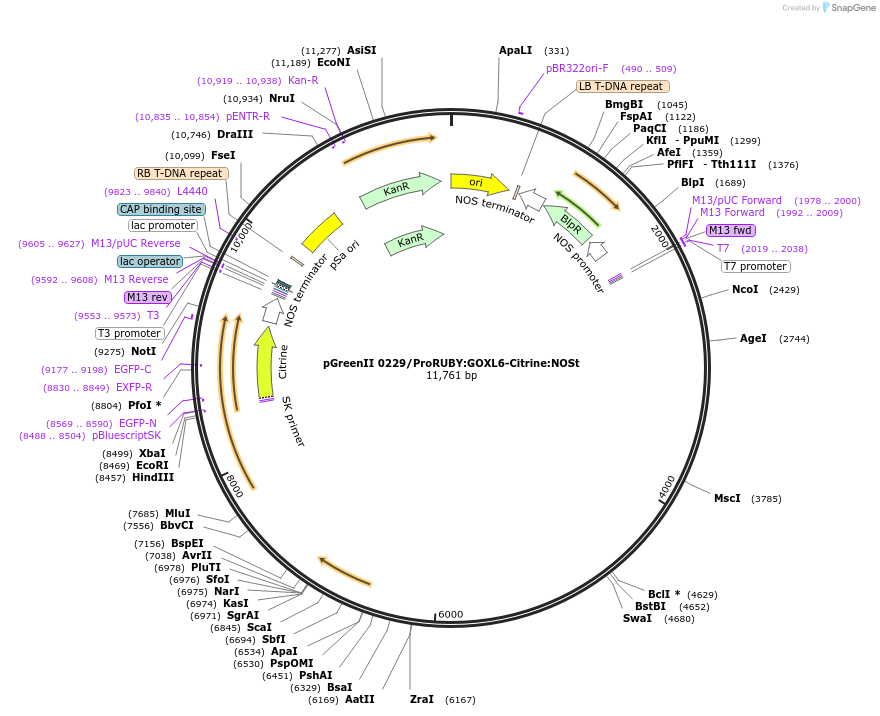
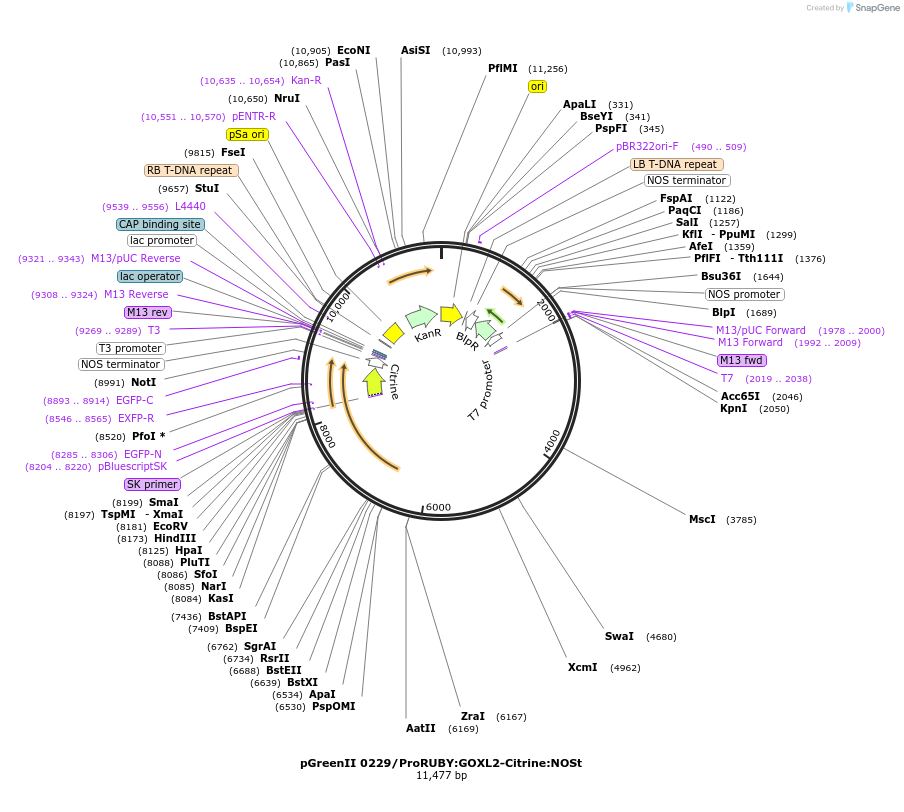
pGreenII 0229/ProRUBY:GOXL5-Citrine:NOSt
Plasmid#175567PurposepSOUP helper plasmid needed for Agrobacterium-mediated plant transformation - Expression of GOXL genes tagged with Citrine under RUBY promoter for transgene complementation of ruby-5DepositorInsertGOXL5
TagsCitrineExpressionPlantPromoterRUBYAvailable SinceOct. 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
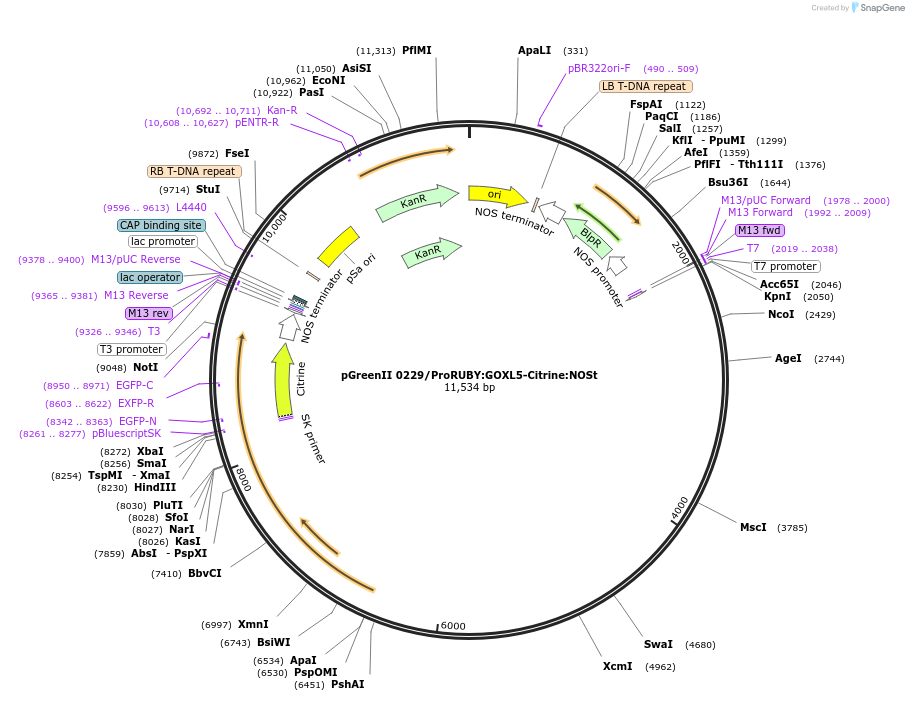
pGreenII 0229/ProRUBY:GOXL4-Citrine:NOSt
Plasmid#175566PurposepSOUP helper plasmid needed for Agrobacterium-mediated plant transformation - Expression of GOXL genes tagged with Citrine under RUBY promoter for transgene complementation of ruby-4DepositorInsertGOXL4
TagsCitrineExpressionPlantPromoterRUBYAvailable SinceOct. 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
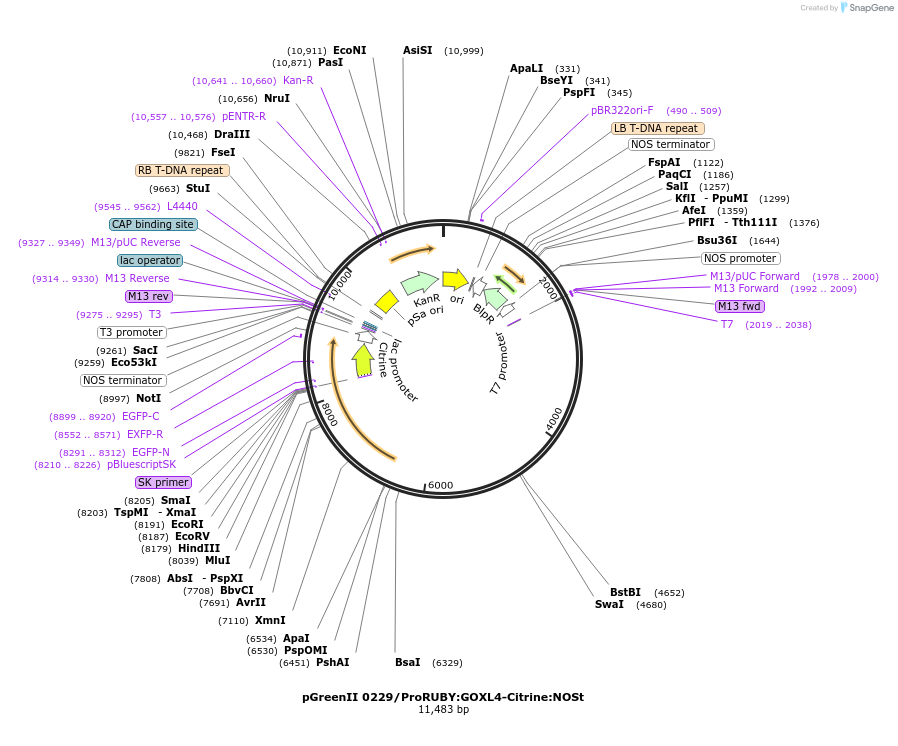
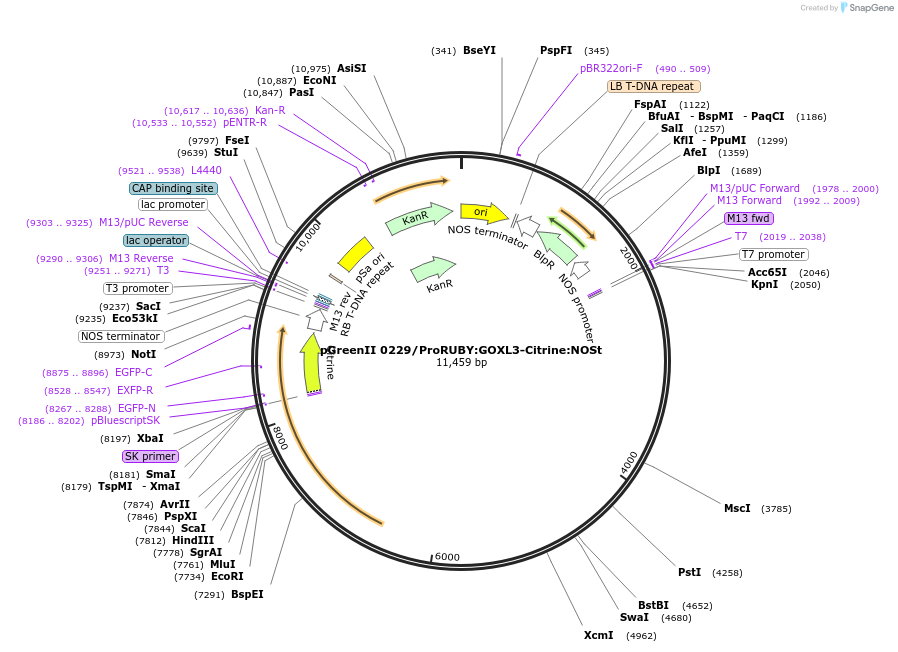
pGreenII 0229/ProRUBY:GOXL3-Citrine:NOSt
Plasmid#175565PurposepSOUP helper plasmid needed for Agrobacterium-mediated plant transformation - Expression of GOXL genes tagged with Citrine under RUBY promoter for transgene complementation of ruby-3DepositorInsertGOXL3
TagsCitrineExpressionPlantPromoterRUBYAvailable SinceOct. 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGreenII 0229/ProRUBY:GOXL2-Citrine:NOSt
Plasmid#175564PurposepSOUP helper plasmid needed for Agrobacterium-mediated plant transformation - Expression of GOXL genes tagged with Citrine under RUBY promoter for transgene complementation of ruby-2DepositorInsertGOXL2
TagsCitrineExpressionPlantPromoterRUBYAvailable SinceOct. 19, 2021AvailabilityAcademic Institutions and Nonprofits only -
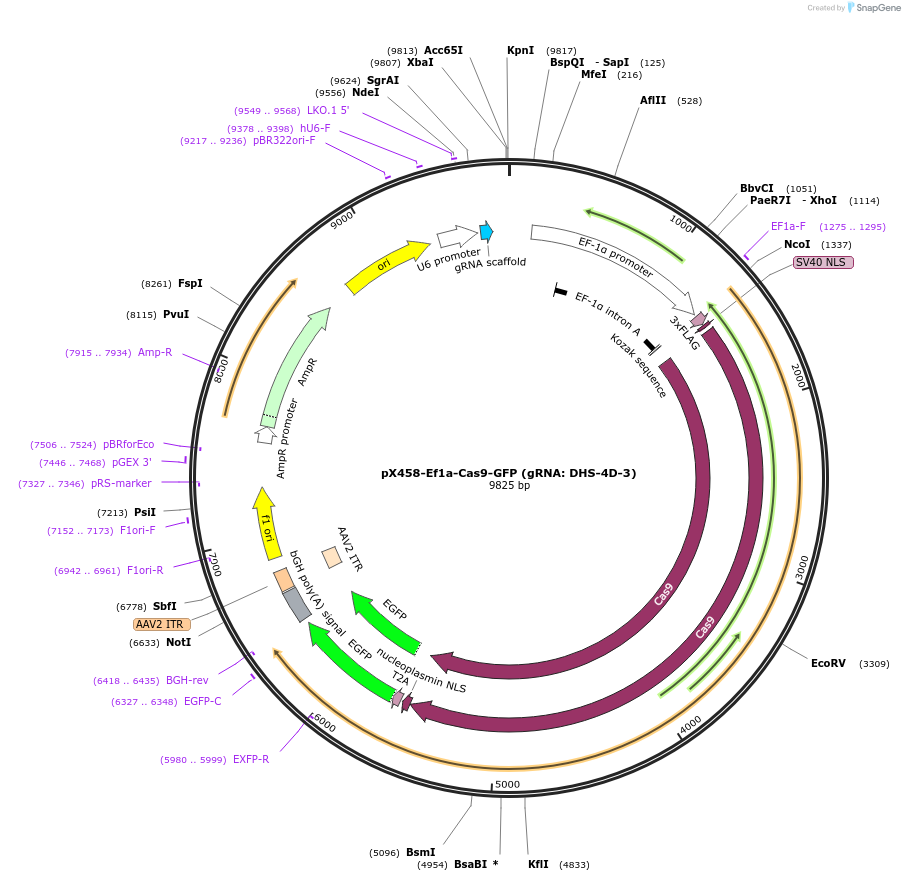
pX458-Ef1a-Cas9-GFP (gRNA: DHS-4D-3)
Plasmid#159664PurposeCRISPR/Cas9 expressing plasmid containing the gRNA targeting an enhancer of mouse Otx2.DepositorInserthSpCas9
UseCRISPRPromoterEf1aAvailable SinceJune 28, 2021AvailabilityAcademic Institutions and Nonprofits only -
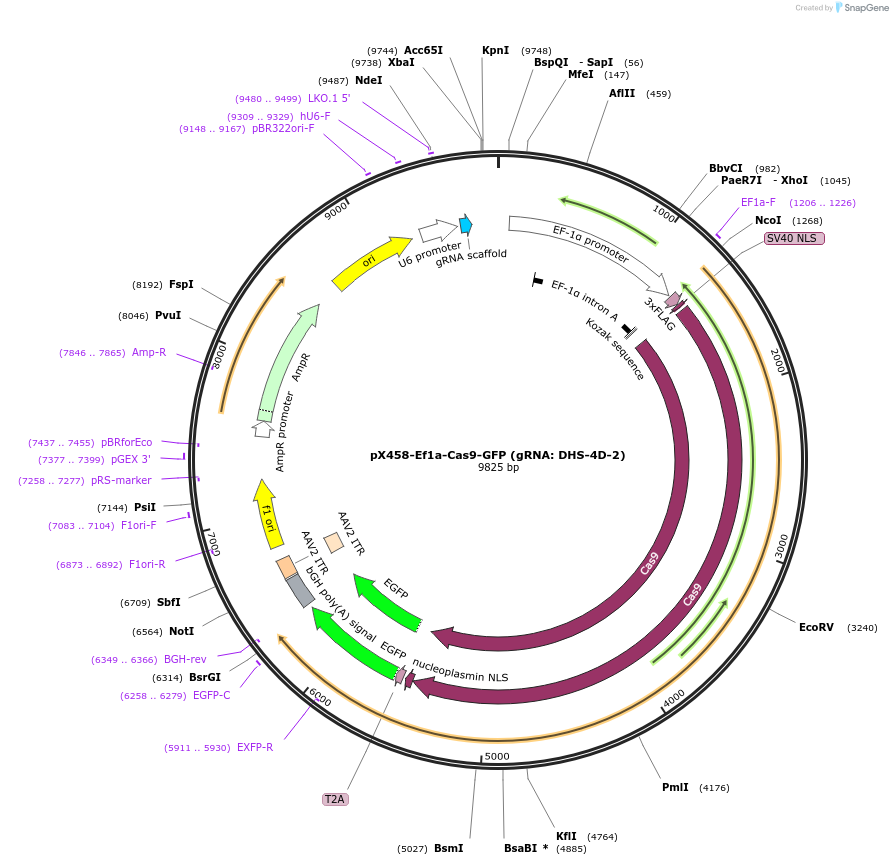
pX458-Ef1a-Cas9-GFP (gRNA: DHS-4D-2)
Plasmid#159663PurposeCRISPR/Cas9 expressing plasmid containing the gRNA targeting an enhancer of mouse Otx2.DepositorInserthSpCas9
UseCRISPRPromoterEf1aAvailable SinceMarch 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
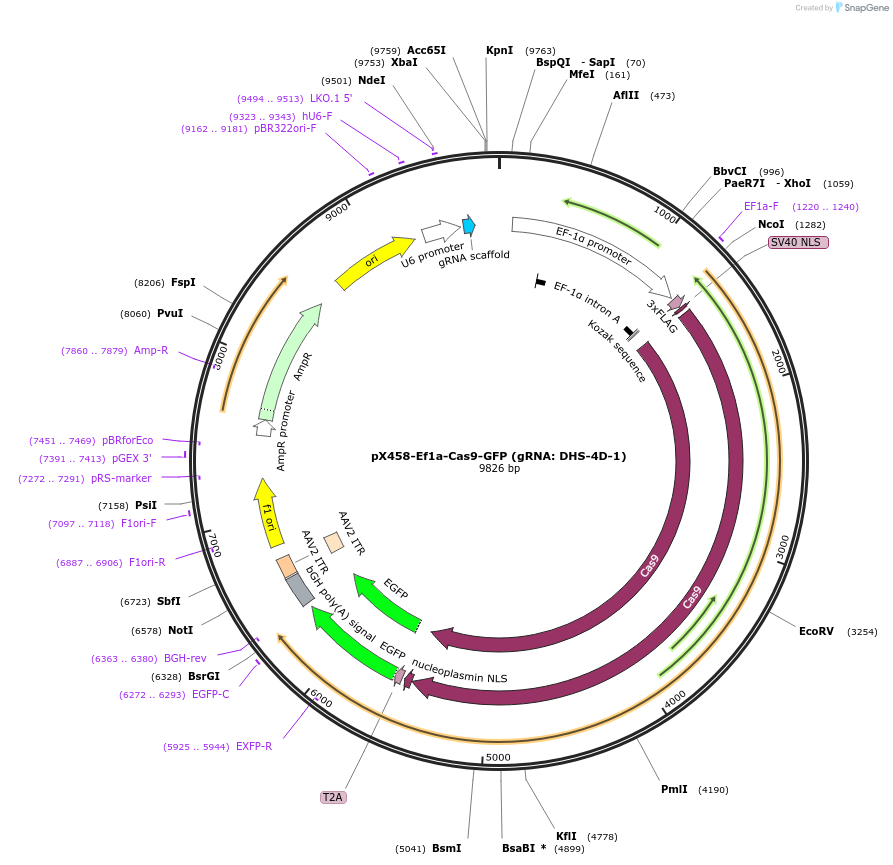
pX458-Ef1a-Cas9-GFP (gRNA: DHS-4D-1)
Plasmid#159662PurposeCRISPR/Cas9 expressing plasmid containing the gRNA targeting an enhancer of mouse Otx2.DepositorInserthSpCas9
UseCRISPRPromoterEf1aAvailable SinceMarch 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
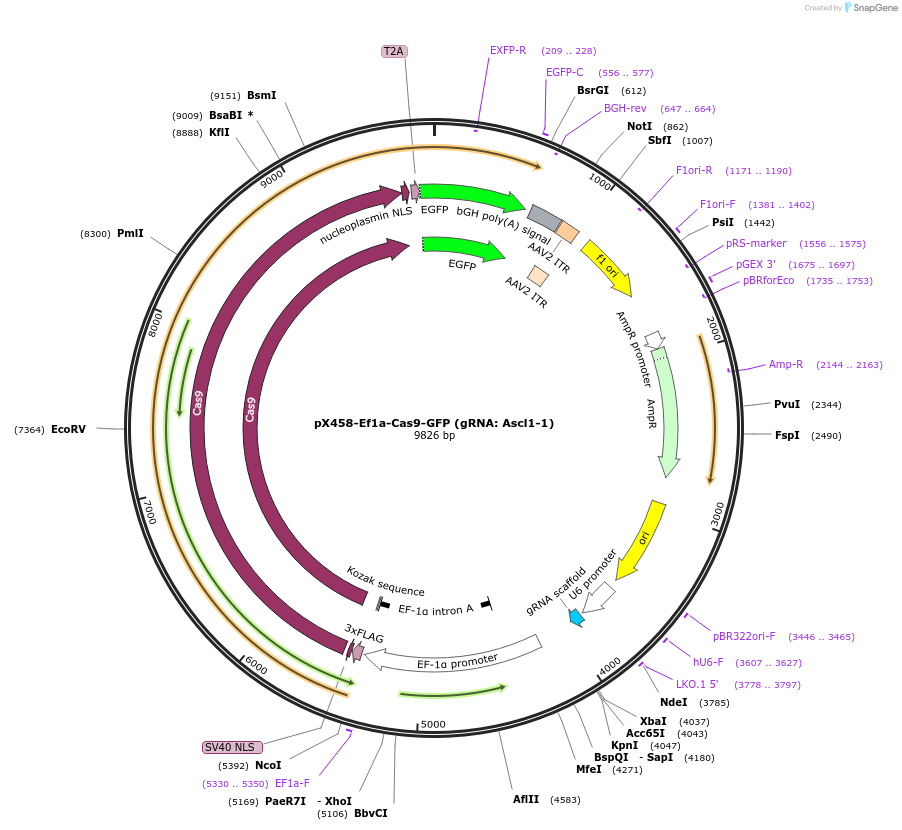
pX458-Ef1a-Cas9-GFP (gRNA: Ascl1-1)
Plasmid#159656PurposeCRISPR/Cas9 expressing plasmid containing the gRNA targeting mouse Ascl1.DepositorInserthSpCas9
UseCRISPRPromoterEf1aAvailable SinceMarch 19, 2021AvailabilityAcademic Institutions and Nonprofits only -
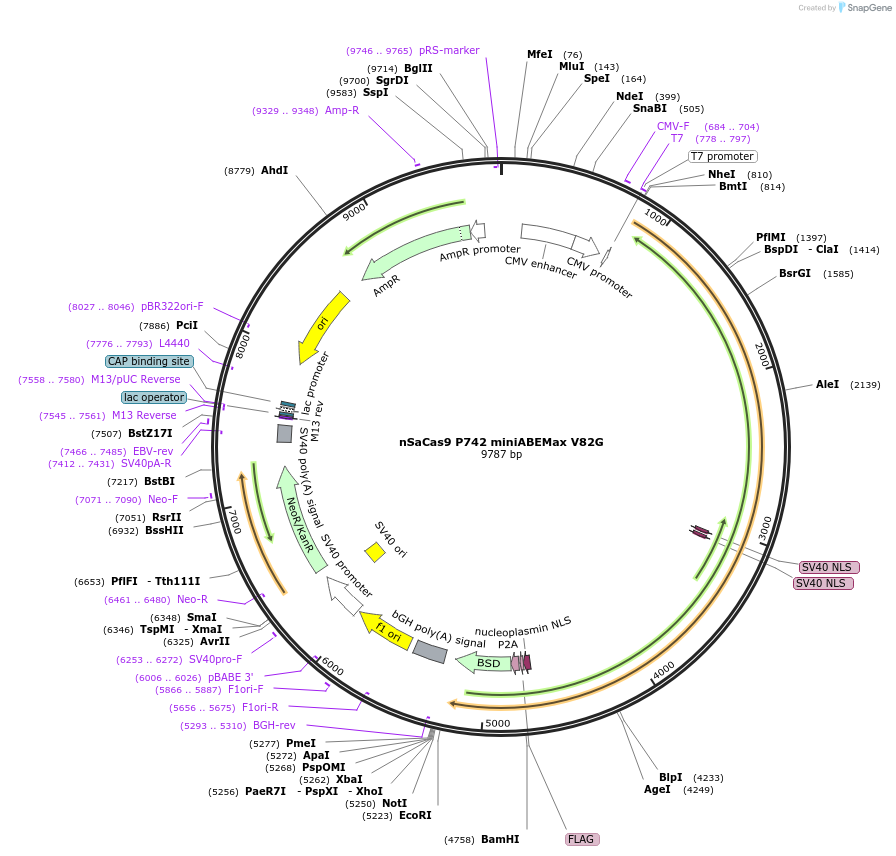
nSaCas9 P742 miniABEMax V82G
Plasmid#135385PurposeExpression of nSaCas9 intradomain insertion of single evolved TadA monomer (V82G) at the specified position (P742 )DepositorInsertnSaCas9 P742 miniABEMax V82G
UseCRISPRExpressionMammalianMutation(V82G)Available SinceOct. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
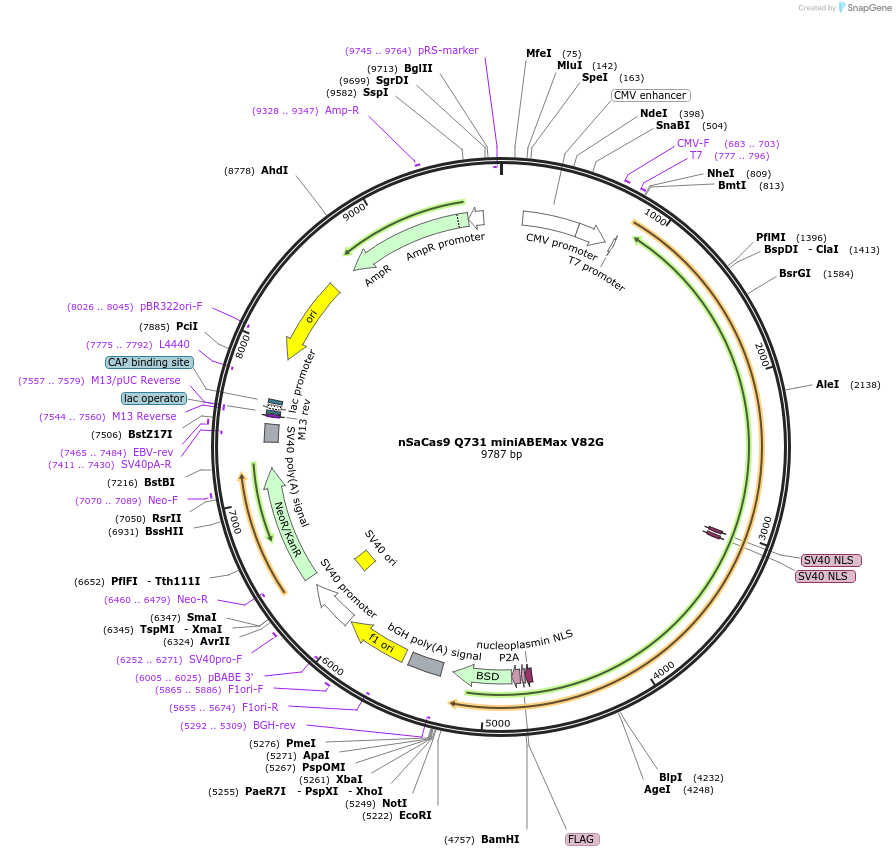
nSaCas9 Q731 miniABEMax V82G
Plasmid#135386PurposeExpression of nSaCas9 intradomain insertion of single evolved TadA monomer (V82G) at the specified position (Q731 )DepositorInsertnSaCas9 Q731 miniABEMax V82G
UseCRISPRExpressionMammalianMutation(V82G)Available SinceOct. 1, 2020AvailabilityAcademic Institutions and Nonprofits only