We narrowed to 17,771 results for: puro
-
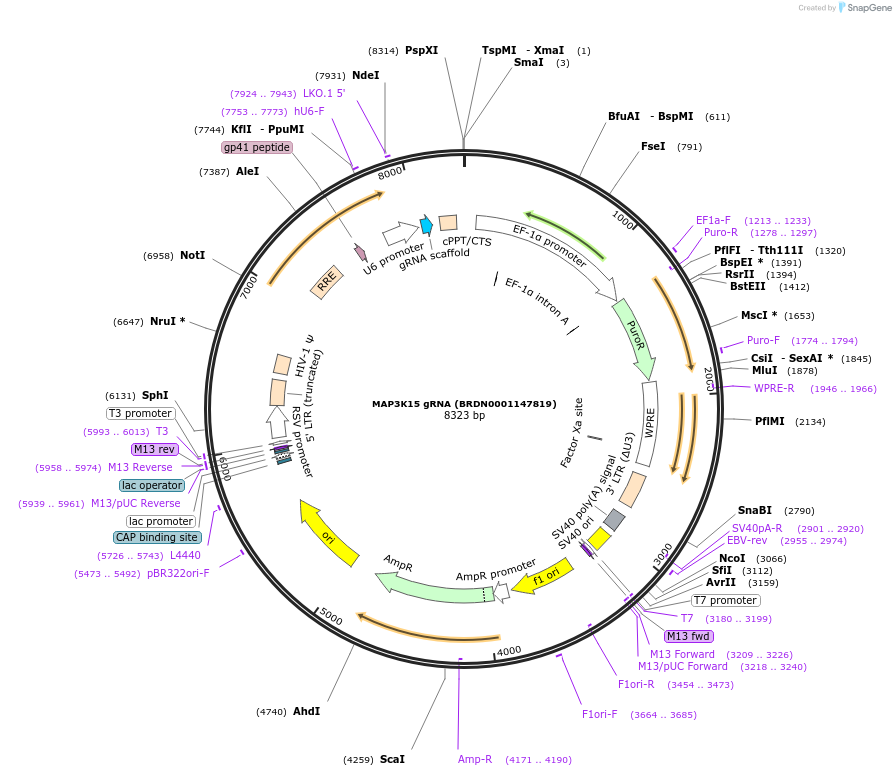
Plasmid#76101Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K15DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only
-
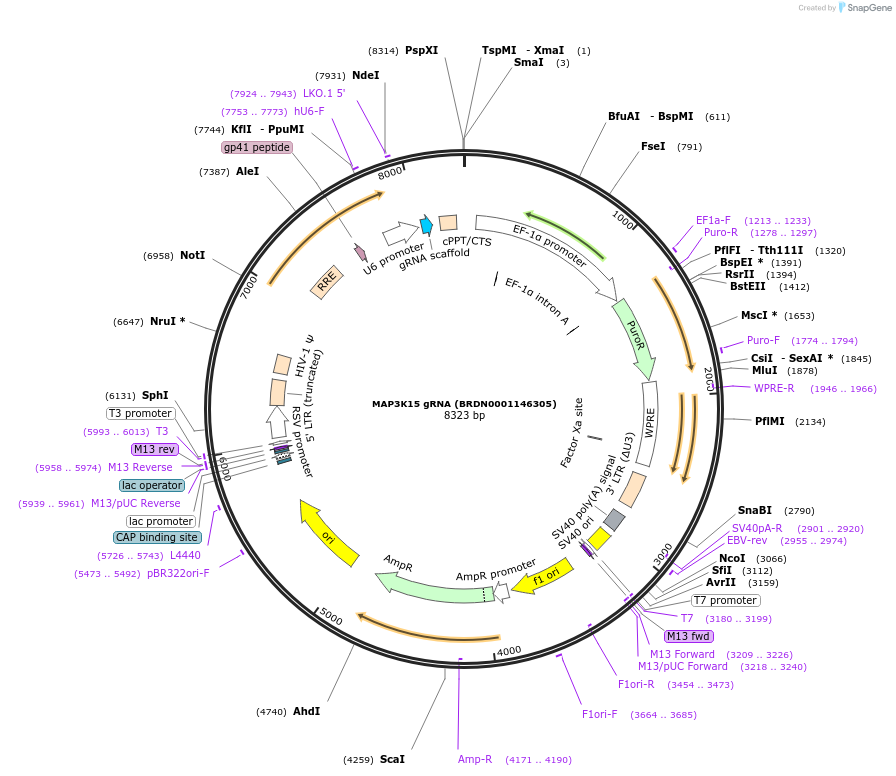
MAP3K15 gRNA (BRDN0001146305)
Plasmid#76102Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K15DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
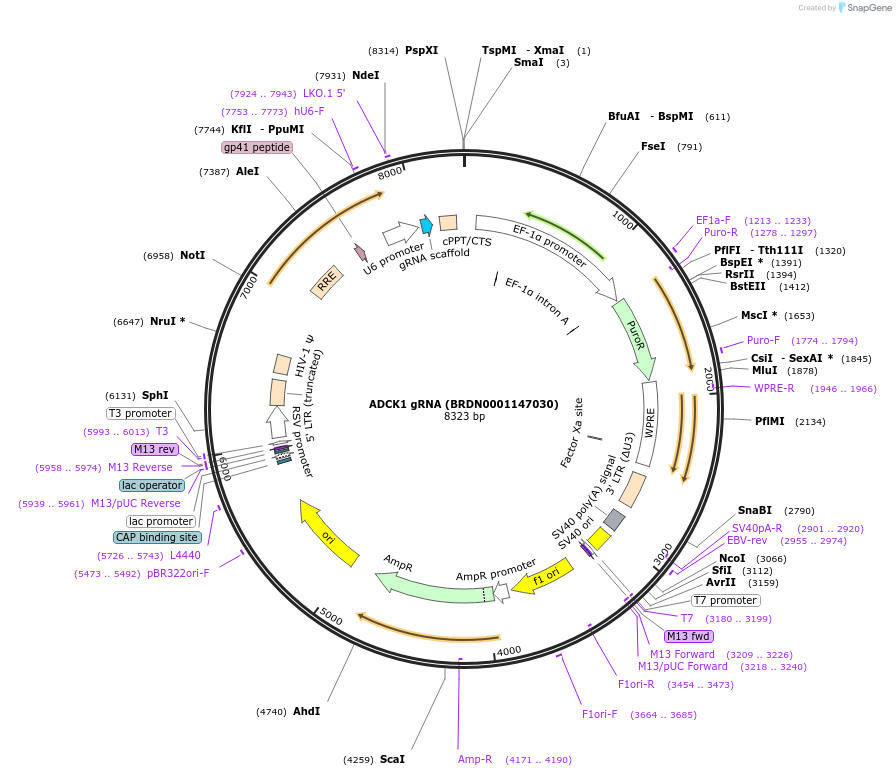
ADCK1 gRNA (BRDN0001147030)
Plasmid#76085Purpose3rd generation lentiviral gRNA plasmid targeting human ADCK1DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
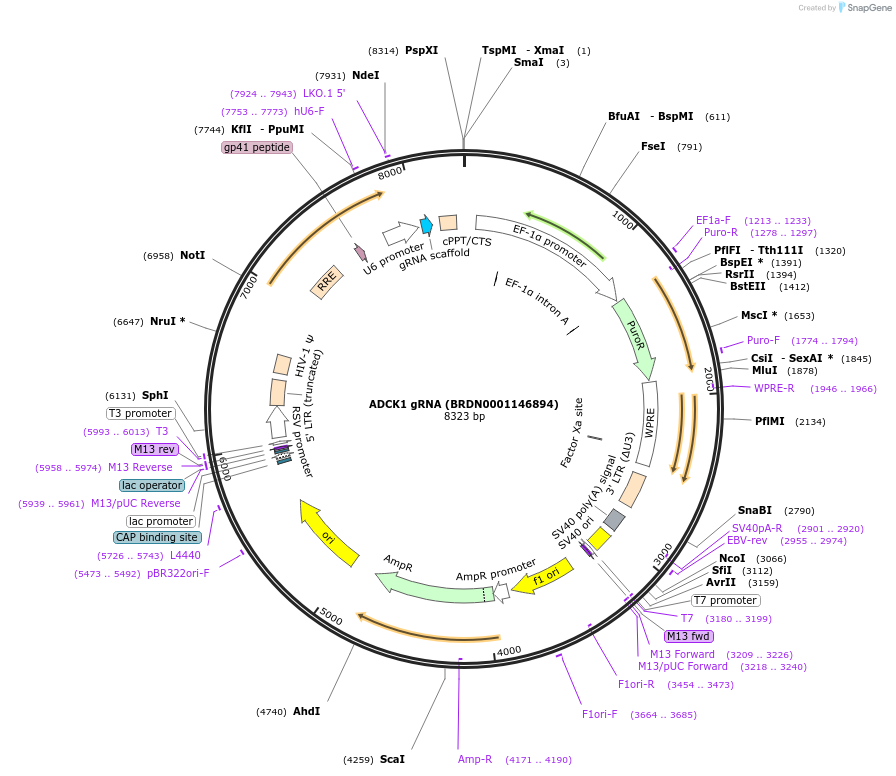
ADCK1 gRNA (BRDN0001146894)
Plasmid#76086Purpose3rd generation lentiviral gRNA plasmid targeting human ADCK1DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
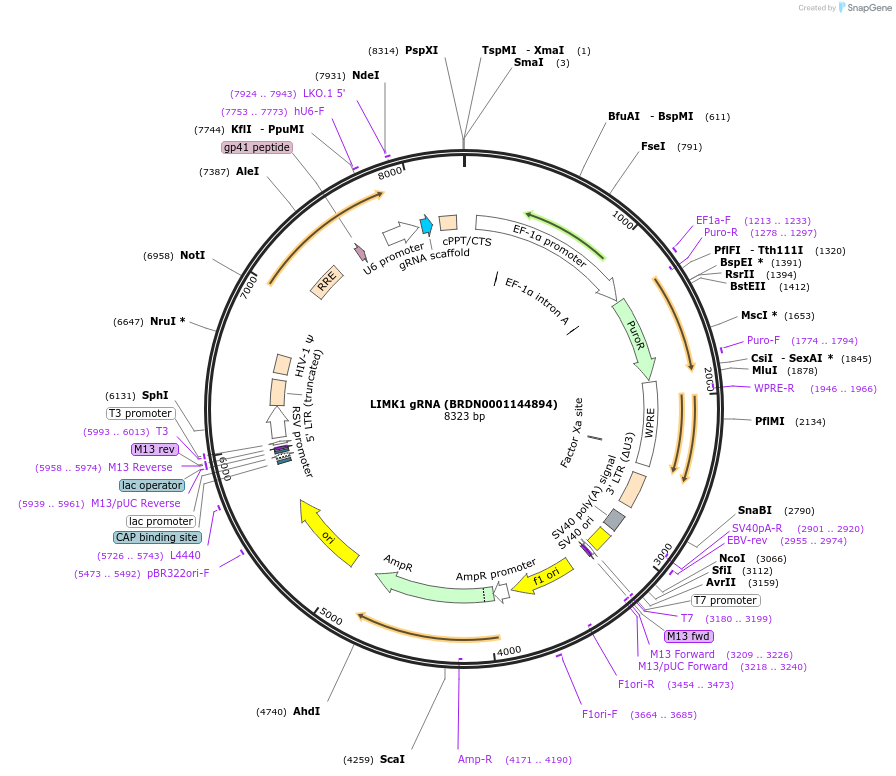
LIMK1 gRNA (BRDN0001144894)
Plasmid#76047Purpose3rd generation lentiviral gRNA plasmid targeting human LIMK1DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
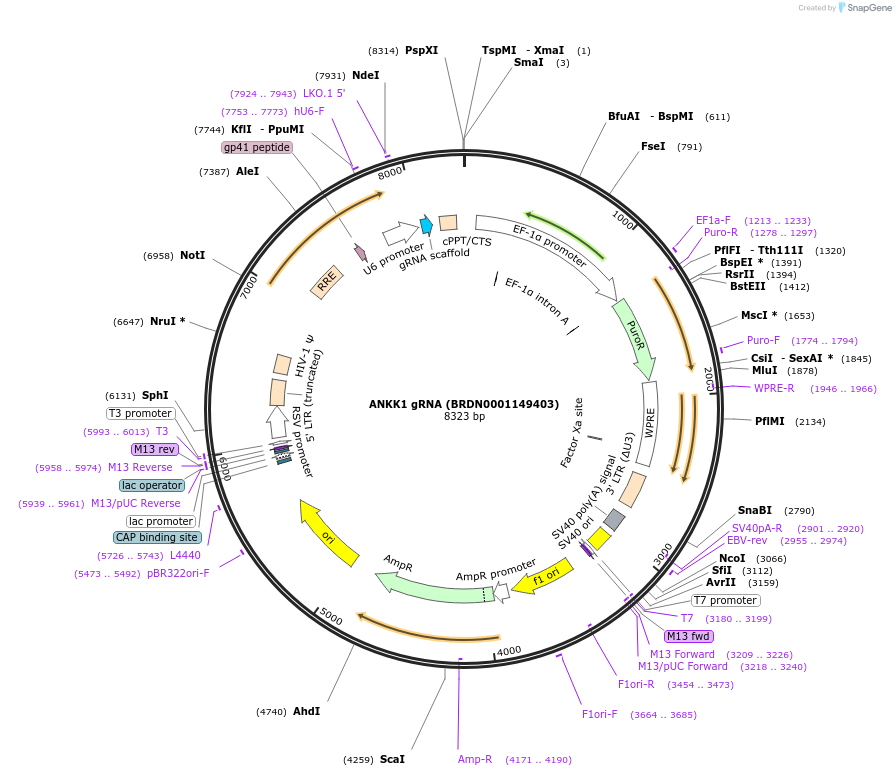
ANKK1 gRNA (BRDN0001149403)
Plasmid#76036Purpose3rd generation lentiviral gRNA plasmid targeting human ANKK1DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
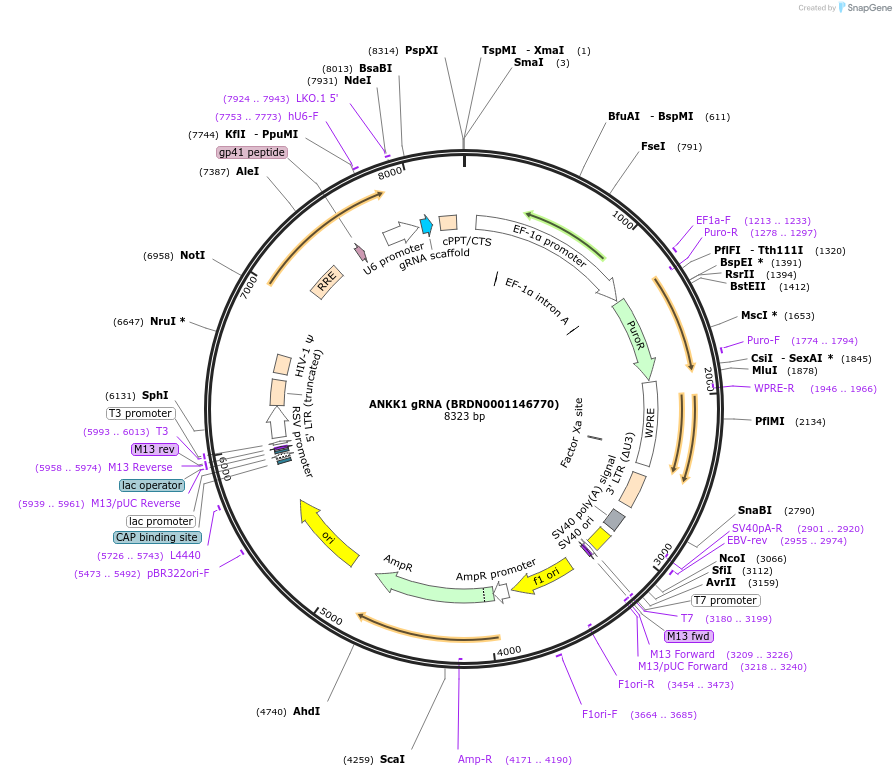
ANKK1 gRNA (BRDN0001146770)
Plasmid#76037Purpose3rd generation lentiviral gRNA plasmid targeting human ANKK1DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
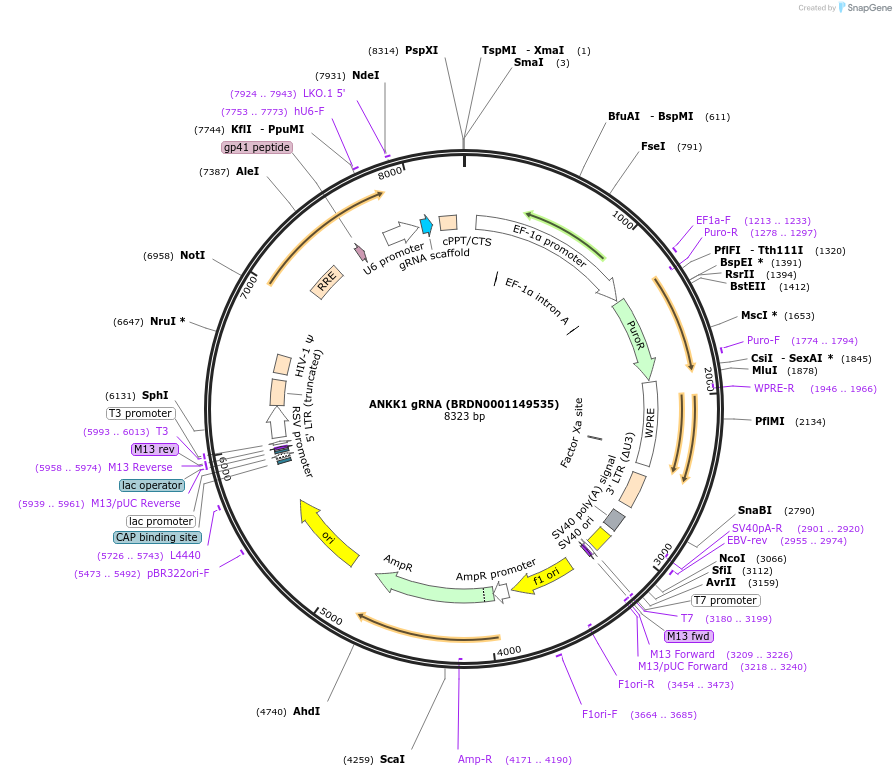
ANKK1 gRNA (BRDN0001149535)
Plasmid#76038Purpose3rd generation lentiviral gRNA plasmid targeting human ANKK1DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
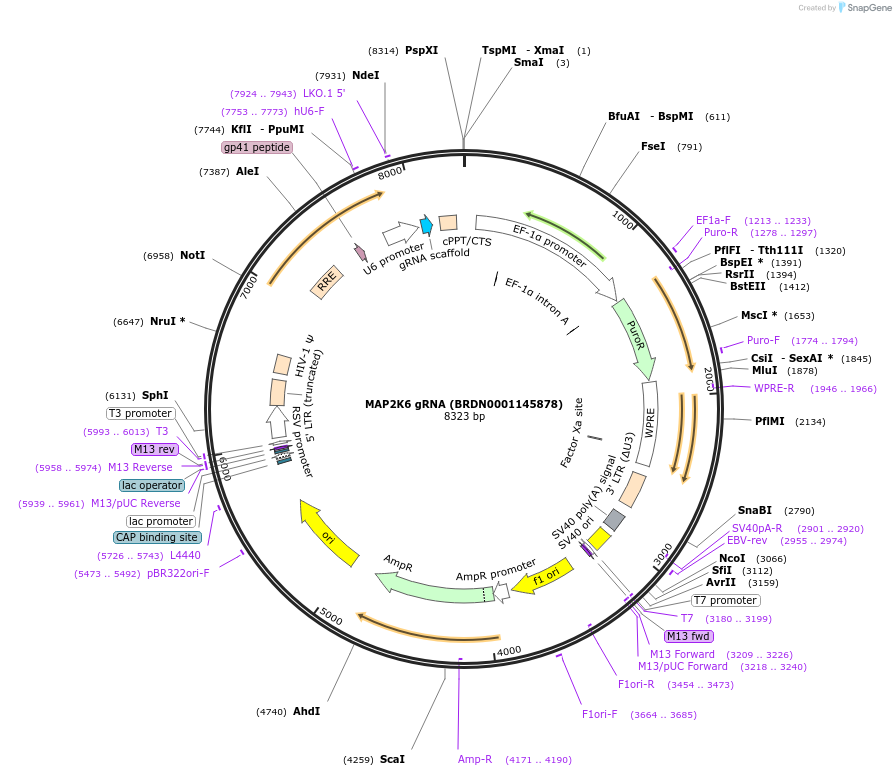
MAP2K6 gRNA (BRDN0001145878)
Plasmid#76039Purpose3rd generation lentiviral gRNA plasmid targeting human MAP2K6DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
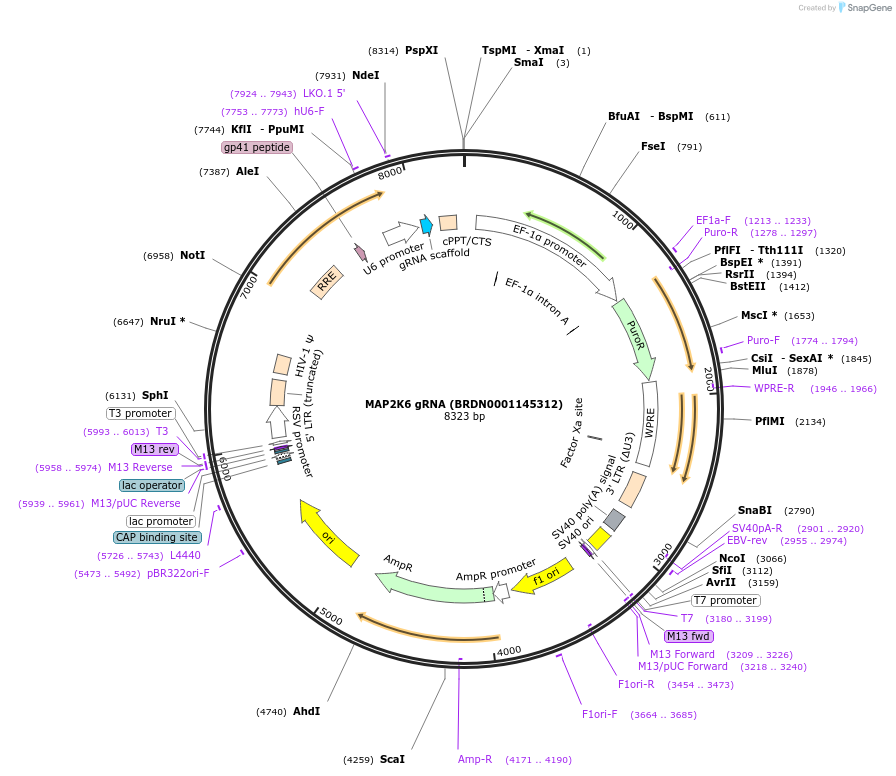
MAP2K6 gRNA (BRDN0001145312)
Plasmid#76040Purpose3rd generation lentiviral gRNA plasmid targeting human MAP2K6DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
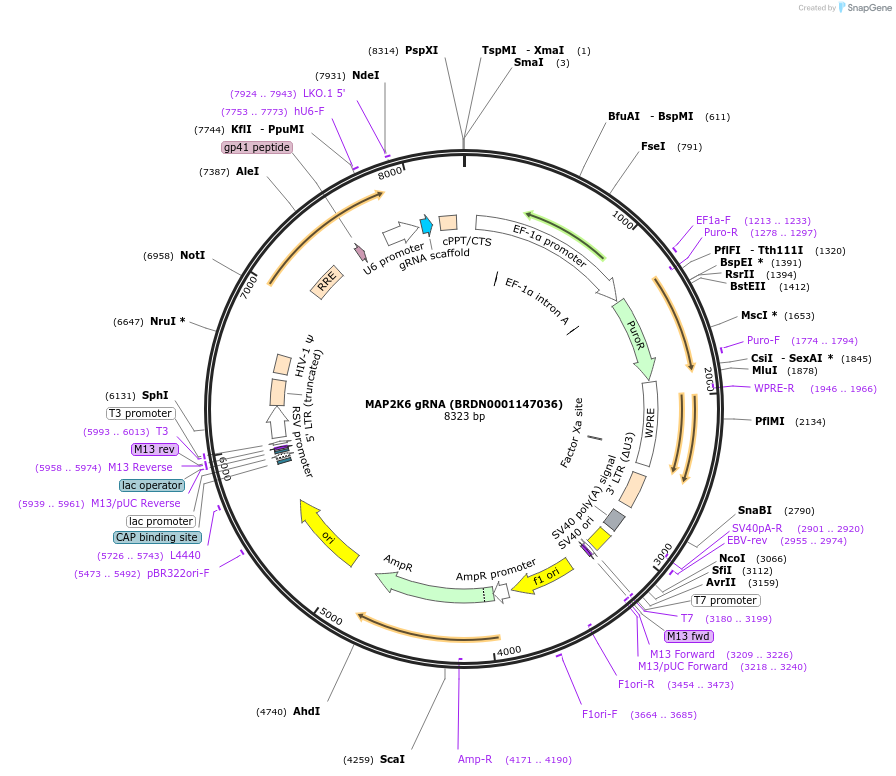
MAP2K6 gRNA (BRDN0001147036)
Plasmid#76041Purpose3rd generation lentiviral gRNA plasmid targeting human MAP2K6DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
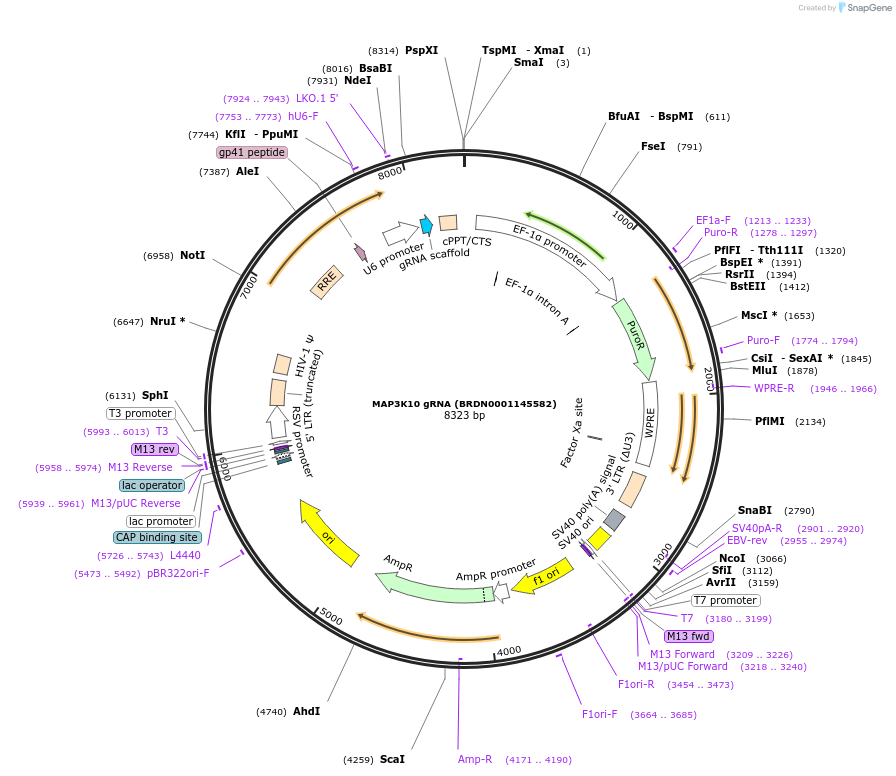
MAP3K10 gRNA (BRDN0001145582)
Plasmid#76006Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K10DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
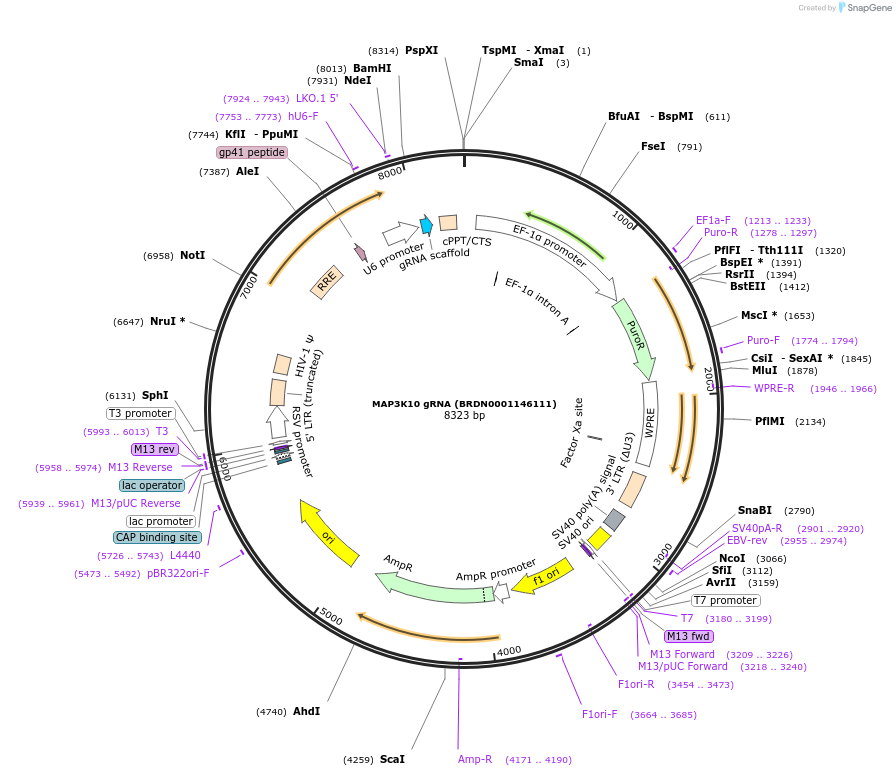
MAP3K10 gRNA (BRDN0001146111)
Plasmid#76007Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K10DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
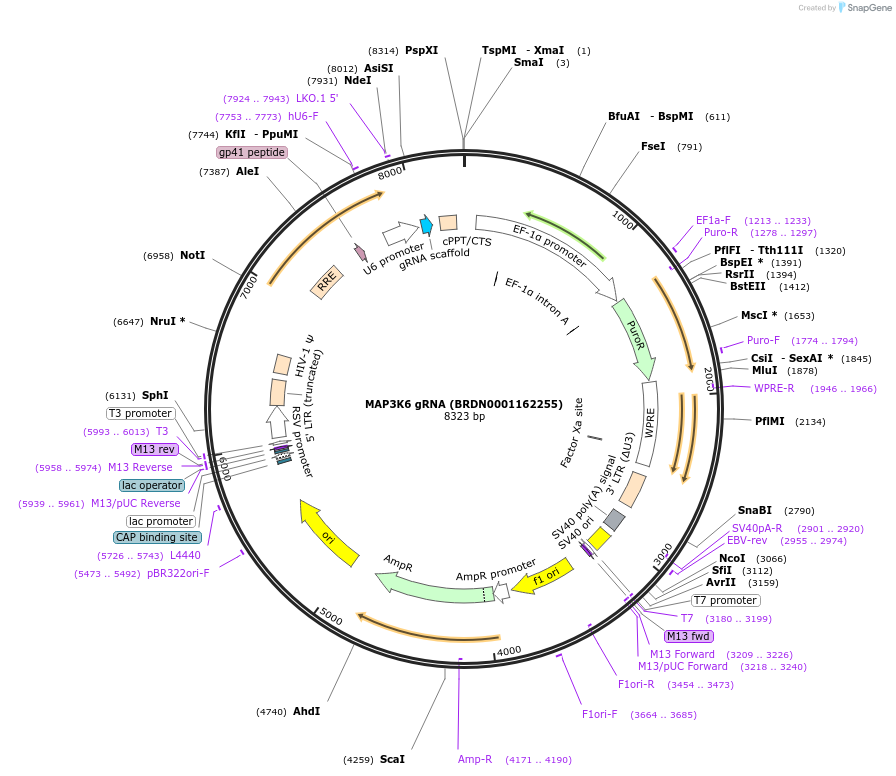
MAP3K6 gRNA (BRDN0001162255)
Plasmid#76012Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K6DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
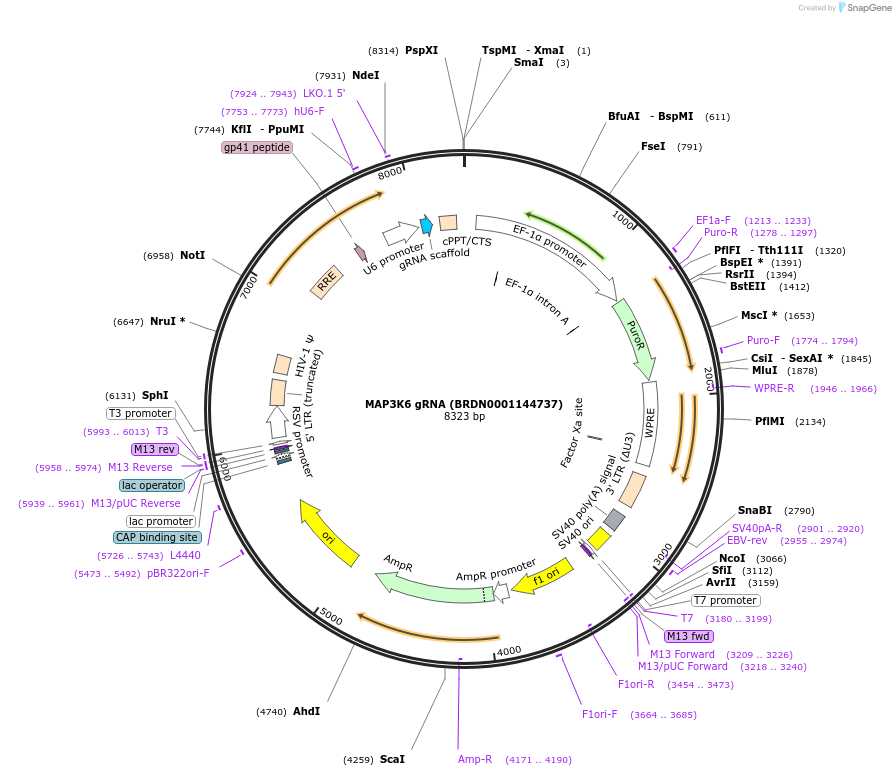
MAP3K6 gRNA (BRDN0001144737)
Plasmid#76013Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K6DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
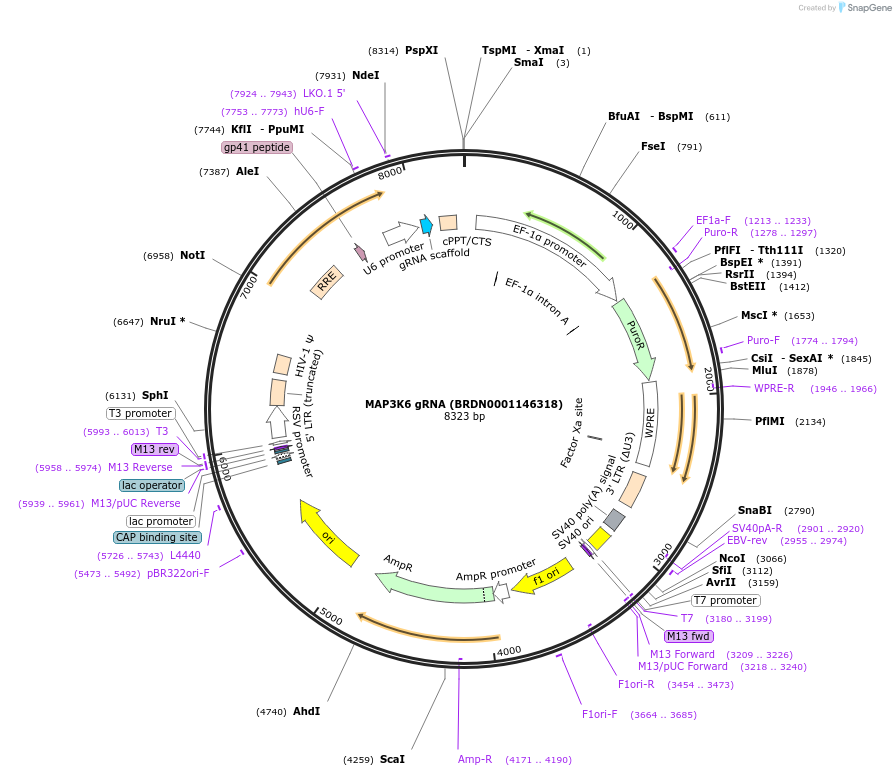
MAP3K6 gRNA (BRDN0001146318)
Plasmid#76014Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K6DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
MAP3K6 gRNA (BRDN0001162229)
Plasmid#76015Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K6DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
MAP3K3 gRNA (BRDN0001149355)
Plasmid#76017Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K3DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
MAP3K3 gRNA (BRDN0001147001)
Plasmid#76018Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K3DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
DAPK2 gRNA (BRDN0001145554)
Plasmid#76019Purpose3rd generation lentiviral gRNA plasmid targeting human DAPK2DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
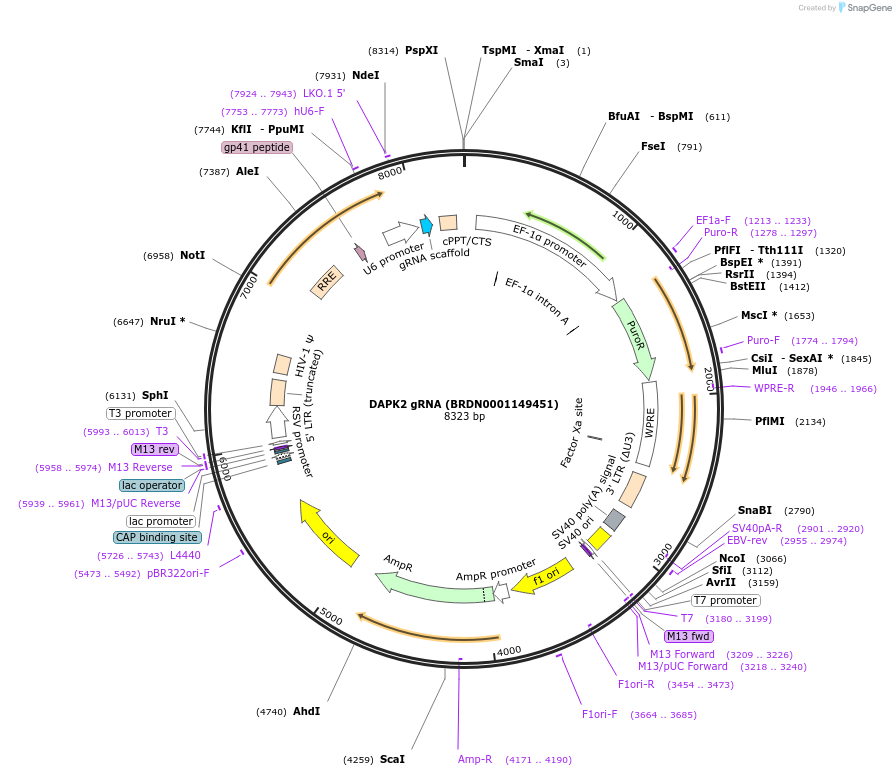
DAPK2 gRNA (BRDN0001149451)
Plasmid#76020Purpose3rd generation lentiviral gRNA plasmid targeting human DAPK2DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
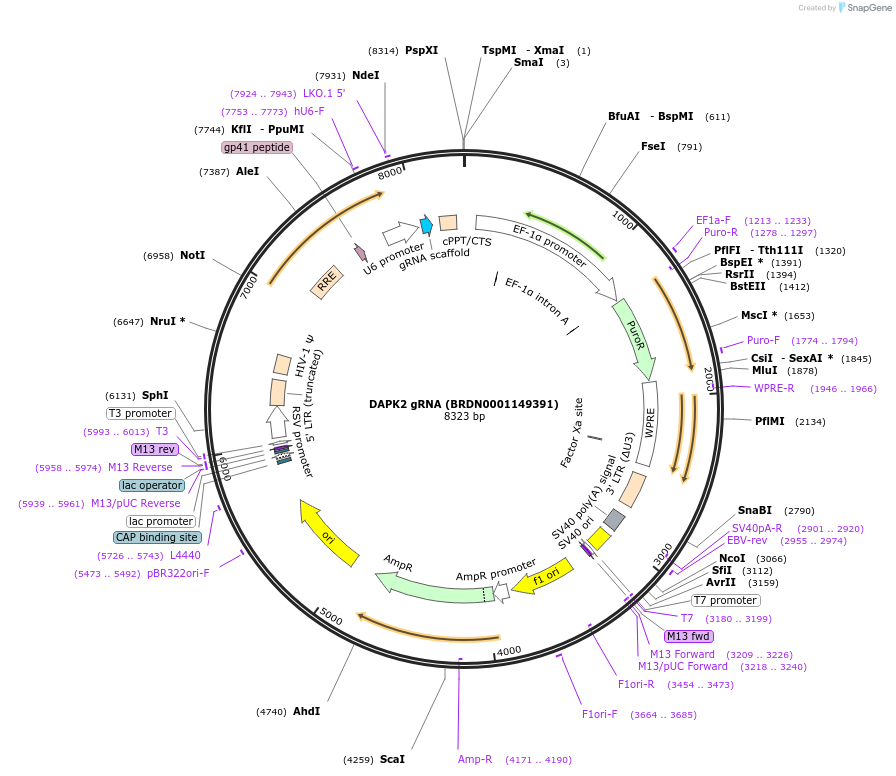
DAPK2 gRNA (BRDN0001149391)
Plasmid#76021Purpose3rd generation lentiviral gRNA plasmid targeting human DAPK2DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
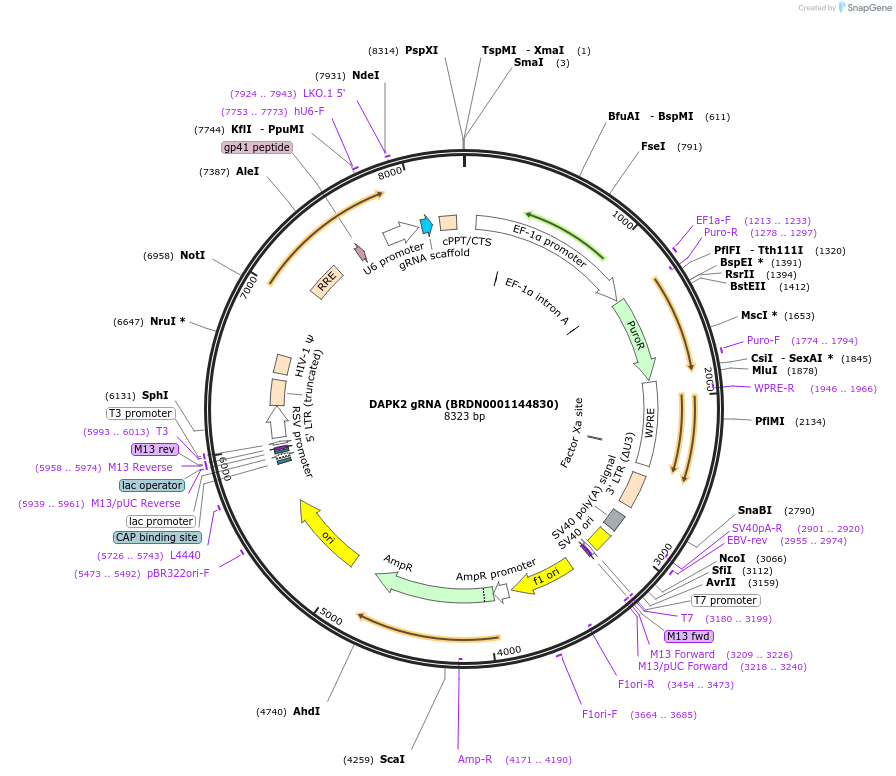
DAPK2 gRNA (BRDN0001144830)
Plasmid#76022Purpose3rd generation lentiviral gRNA plasmid targeting human DAPK2DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
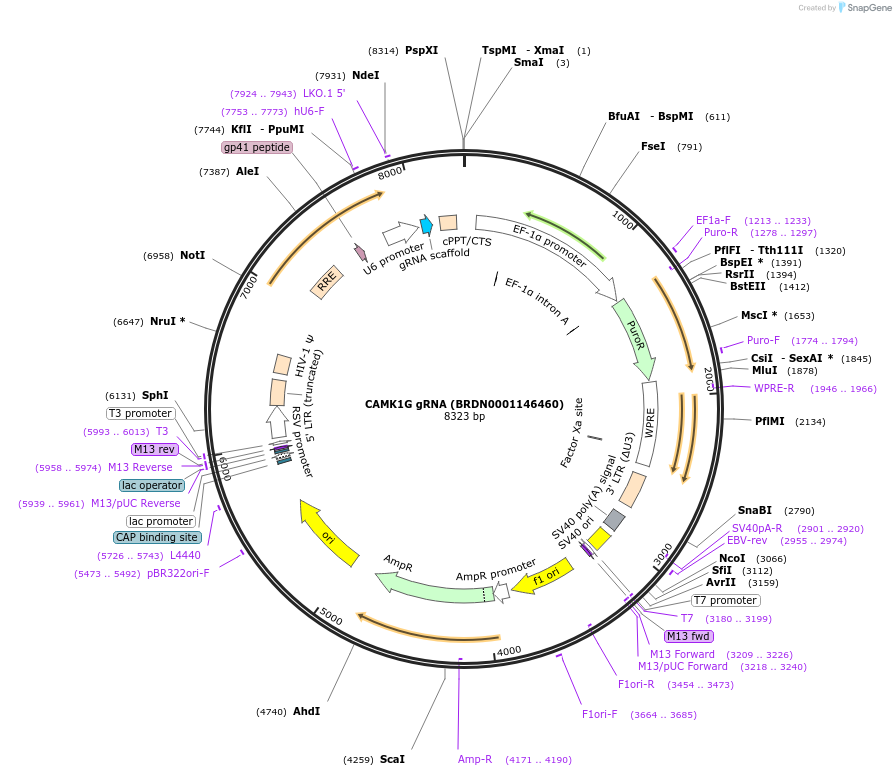
CAMK1G gRNA (BRDN0001146460)
Plasmid#75986Purpose3rd generation lentiviral gRNA plasmid targeting human CAMK1GDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
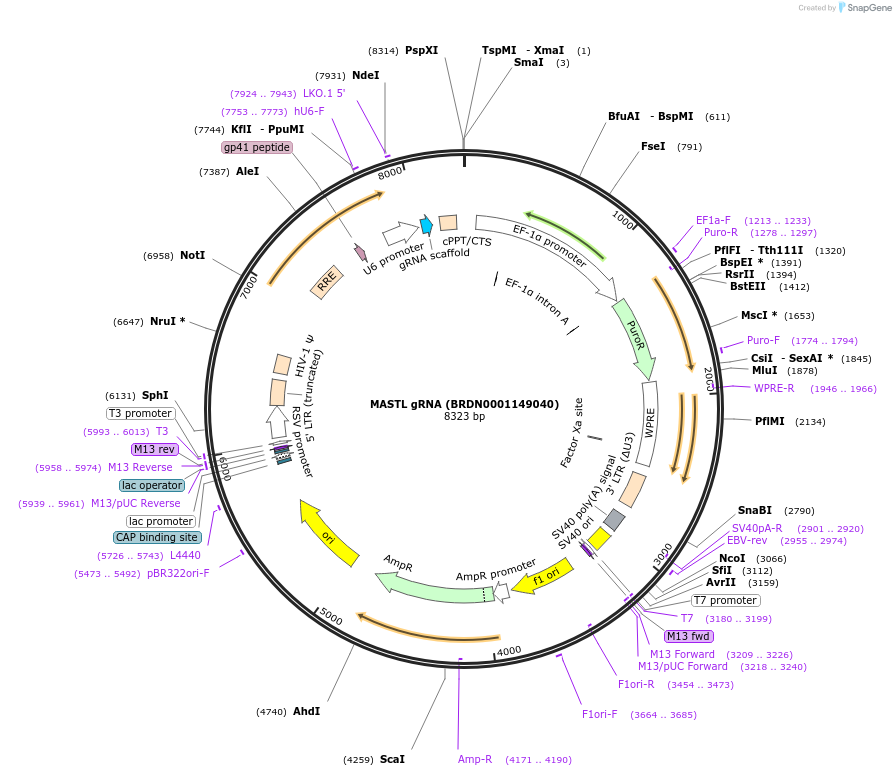
MASTL gRNA (BRDN0001149040)
Plasmid#75988Purpose3rd generation lentiviral gRNA plasmid targeting human MASTLDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
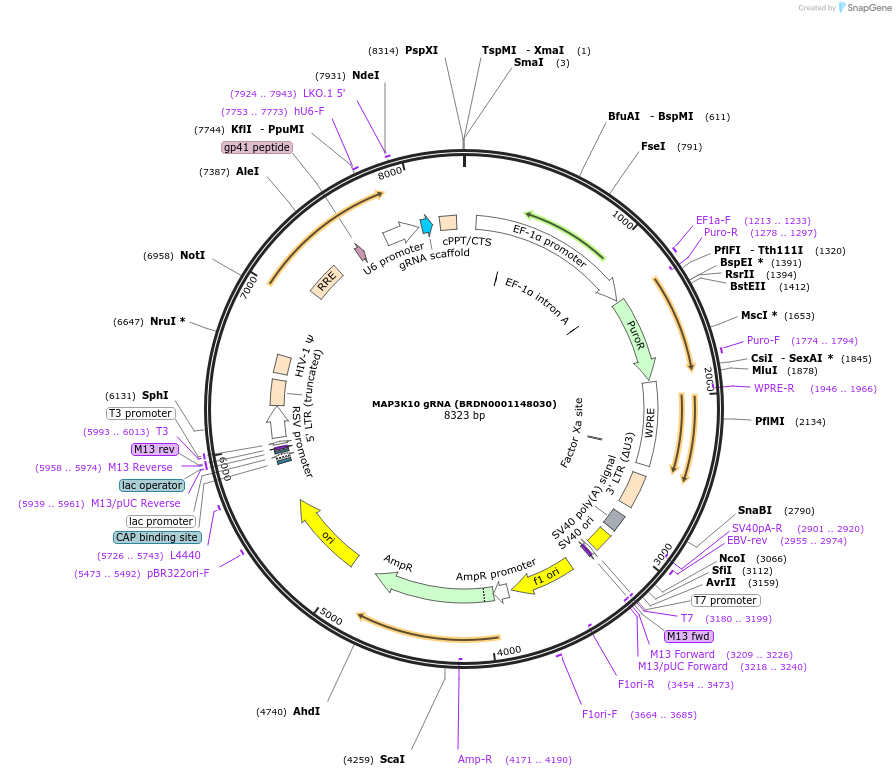
MAP3K10 gRNA (BRDN0001148030)
Plasmid#76005Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K10DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
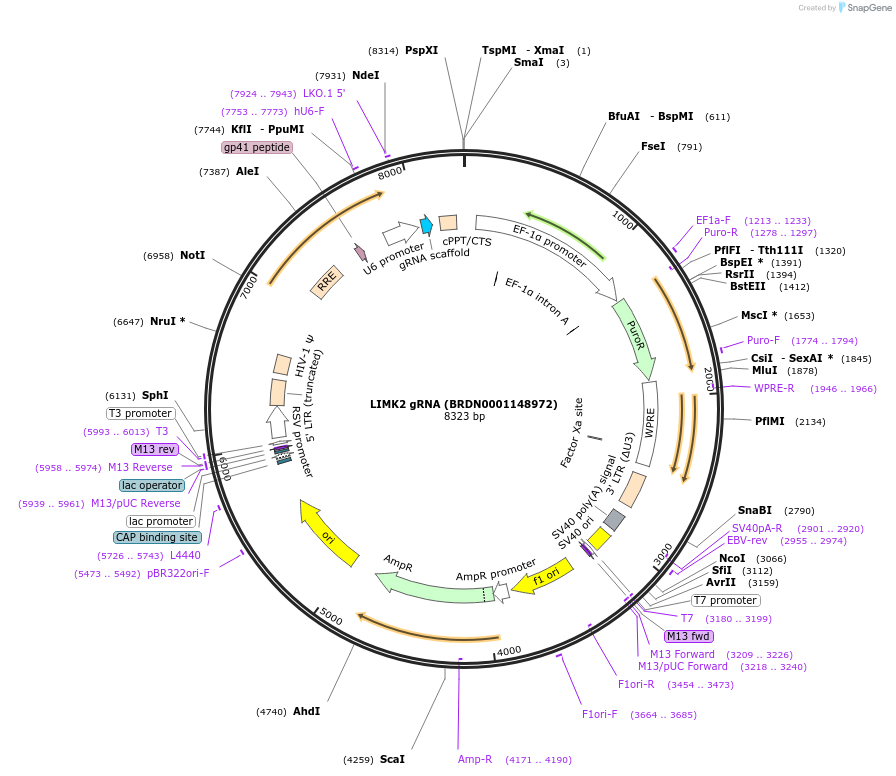
LIMK2 gRNA (BRDN0001148972)
Plasmid#75974Purpose3rd generation lentiviral gRNA plasmid targeting human LIMK2DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
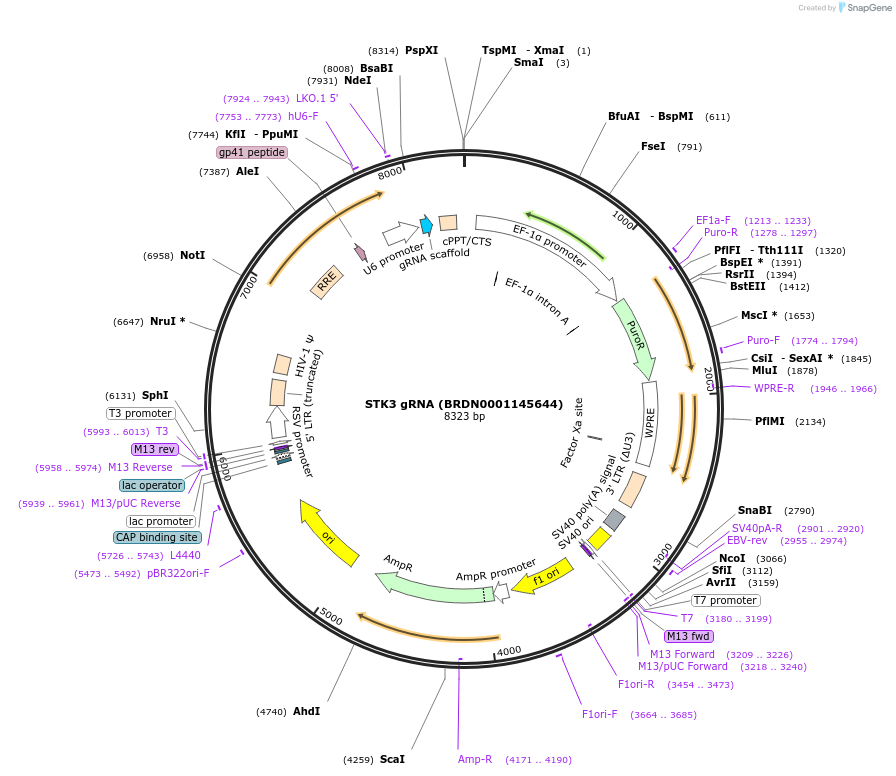
STK3 gRNA (BRDN0001145644)
Plasmid#75976Purpose3rd generation lentiviral gRNA plasmid targeting human STK3DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
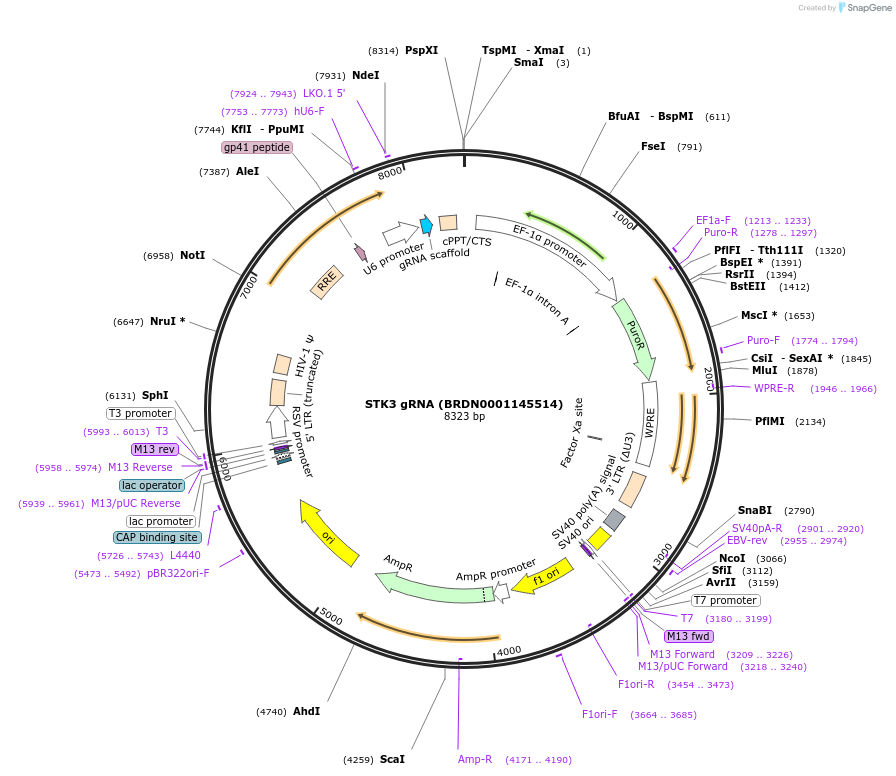
STK3 gRNA (BRDN0001145514)
Plasmid#75977Purpose3rd generation lentiviral gRNA plasmid targeting human STK3DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
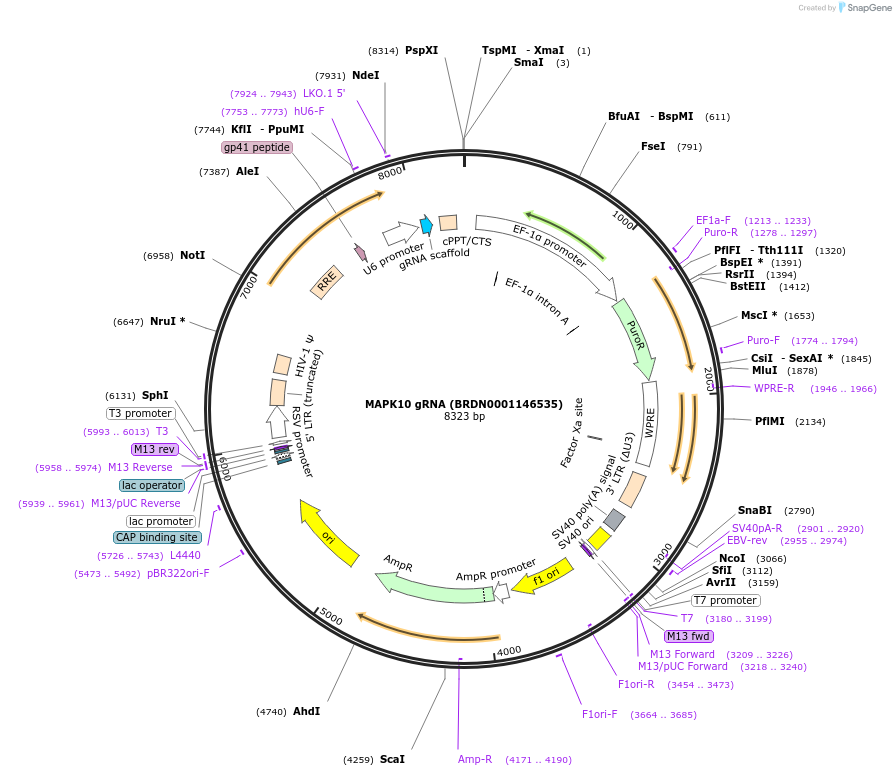
MAPK10 gRNA (BRDN0001146535)
Plasmid#75982Purpose3rd generation lentiviral gRNA plasmid targeting human MAPK10DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
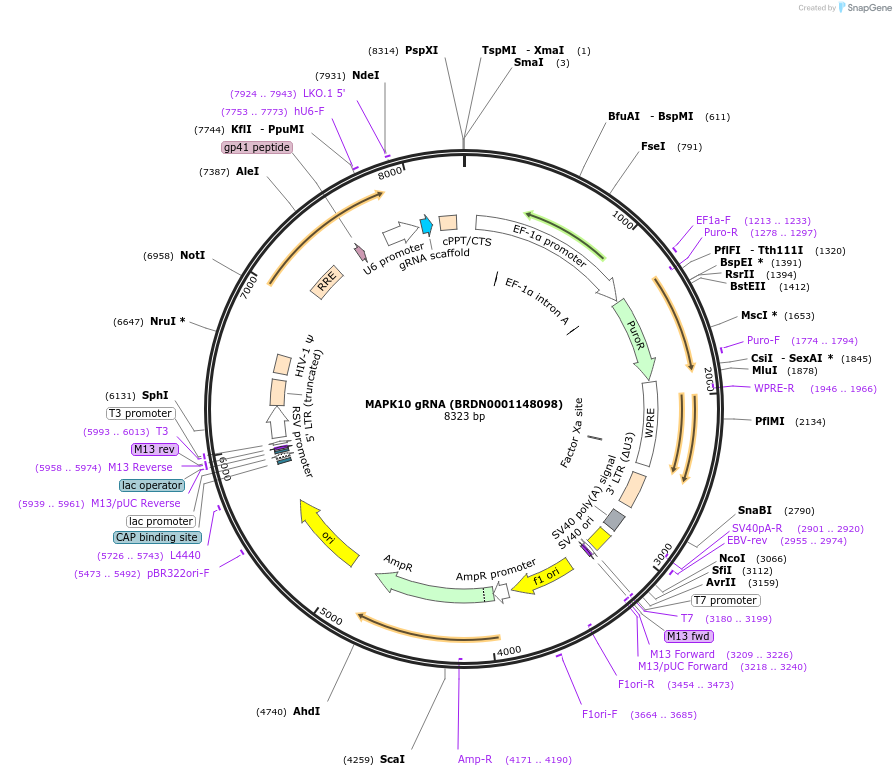
MAPK10 gRNA (BRDN0001148098)
Plasmid#75983Purpose3rd generation lentiviral gRNA plasmid targeting human MAPK10DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
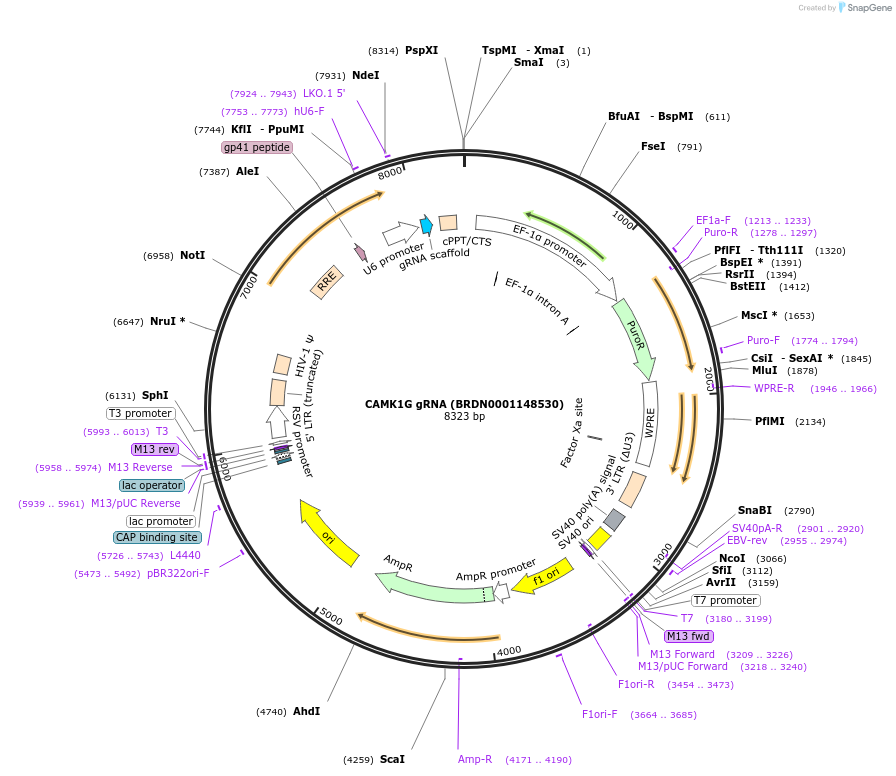
CAMK1G gRNA (BRDN0001148530)
Plasmid#75984Purpose3rd generation lentiviral gRNA plasmid targeting human CAMK1GDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
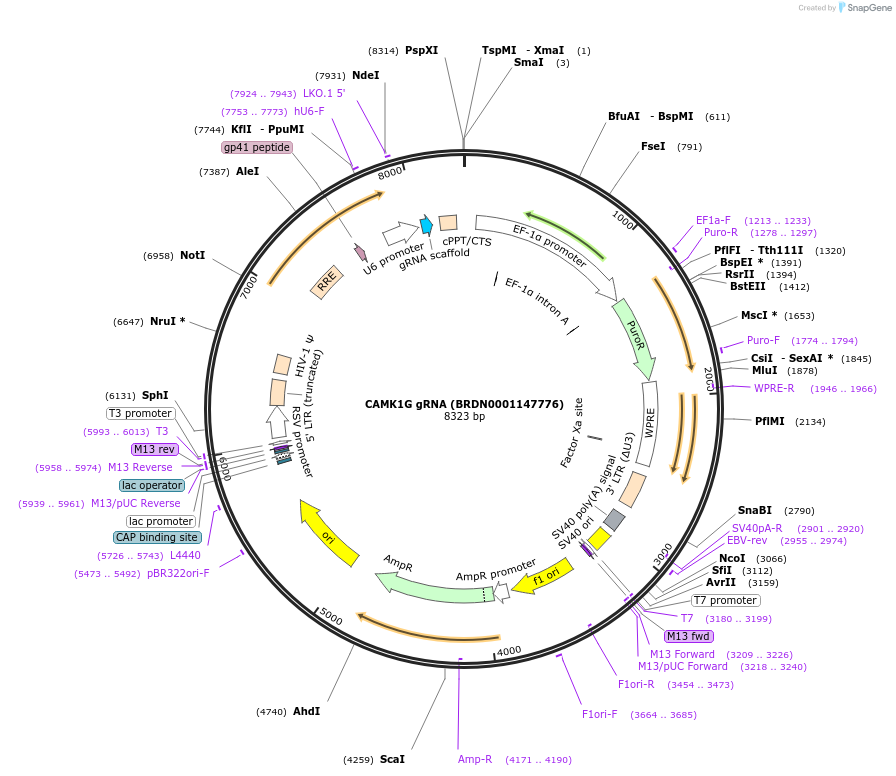
CAMK1G gRNA (BRDN0001147776)
Plasmid#75985Purpose3rd generation lentiviral gRNA plasmid targeting human CAMK1GDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
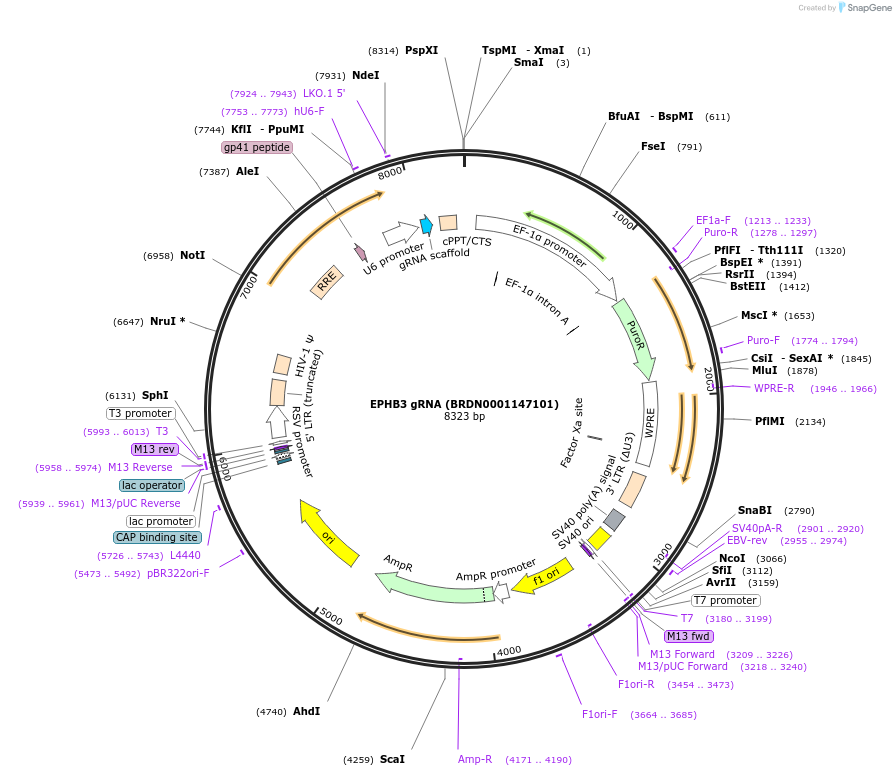
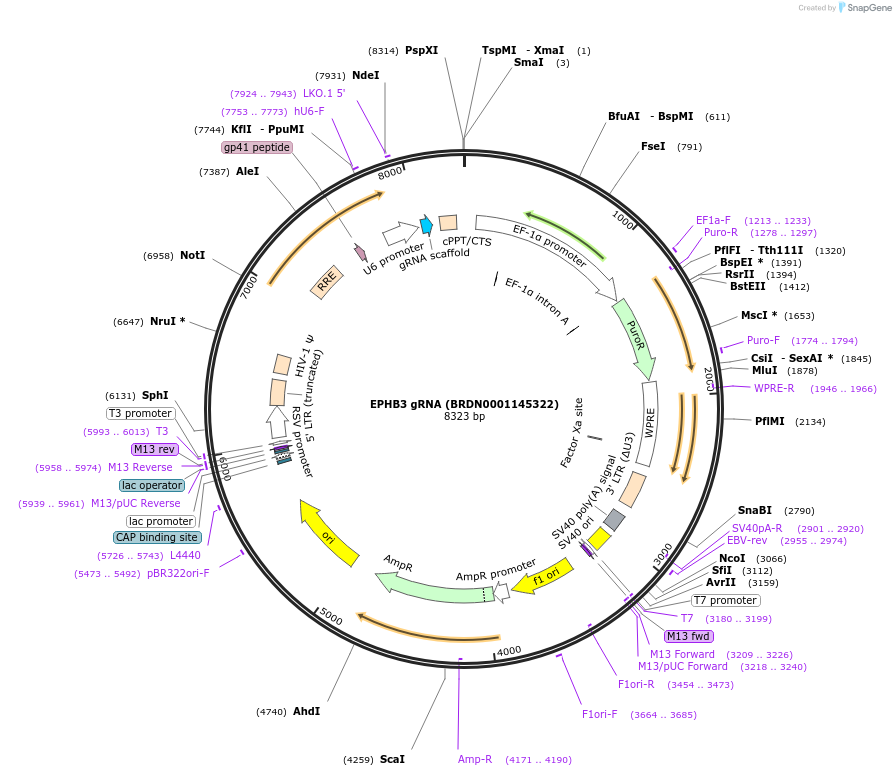
EPHB3 gRNA (BRDN0001147101)
Plasmid#75947Purpose3rd generation lentiviral gRNA plasmid targeting human EPHB3DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
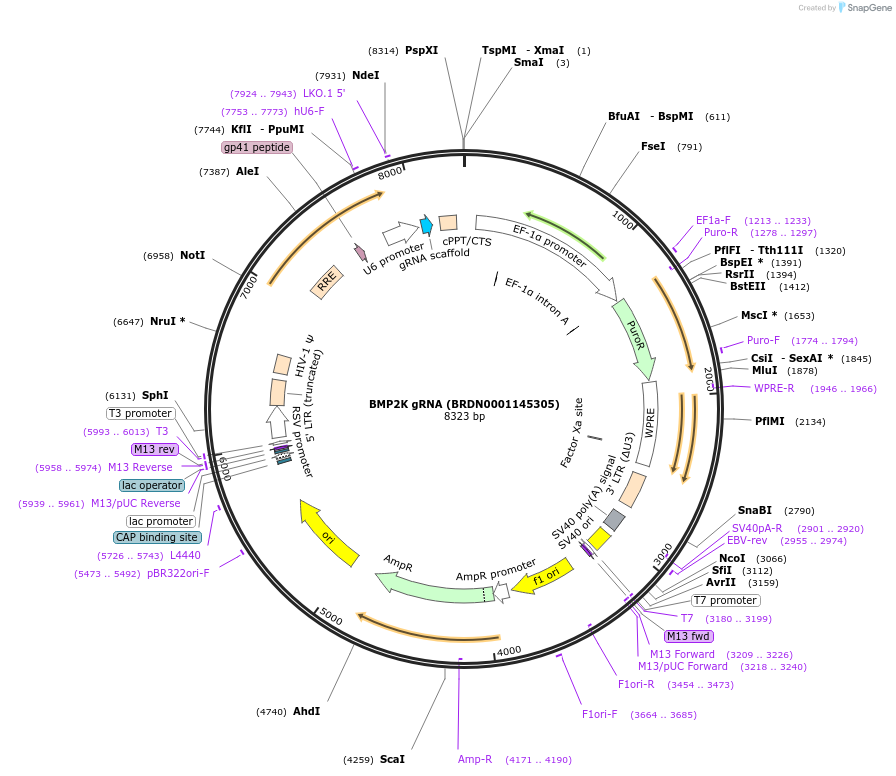
BMP2K gRNA (BRDN0001145305)
Plasmid#75927Purpose3rd generation lentiviral gRNA plasmid targeting human BMP2KDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
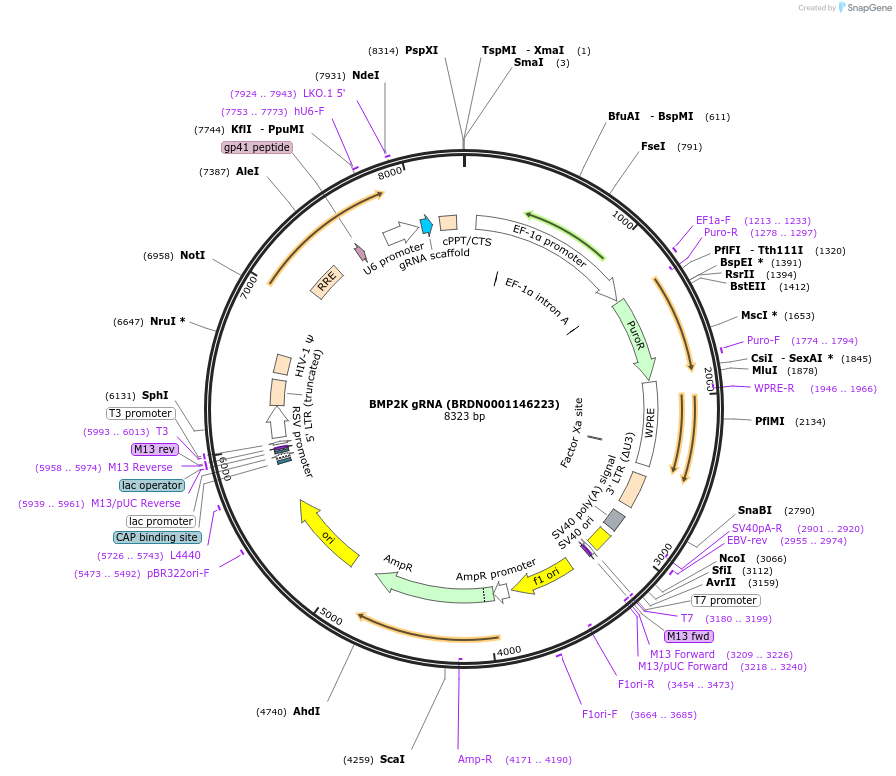
BMP2K gRNA (BRDN0001146223)
Plasmid#75928Purpose3rd generation lentiviral gRNA plasmid targeting human BMP2KDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
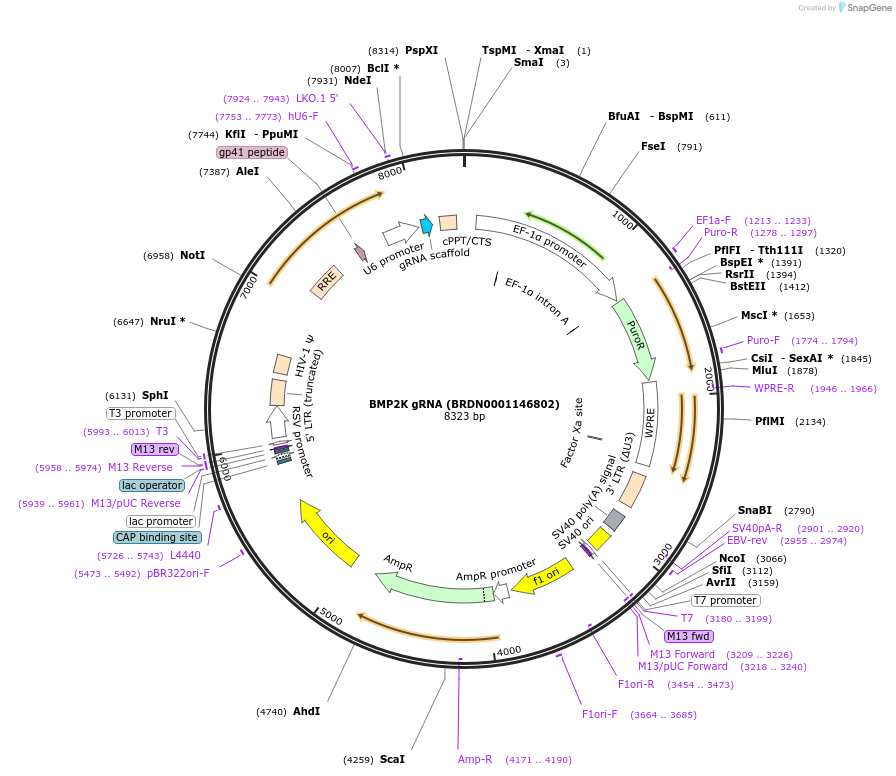
BMP2K gRNA (BRDN0001146802)
Plasmid#75929Purpose3rd generation lentiviral gRNA plasmid targeting human BMP2KDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
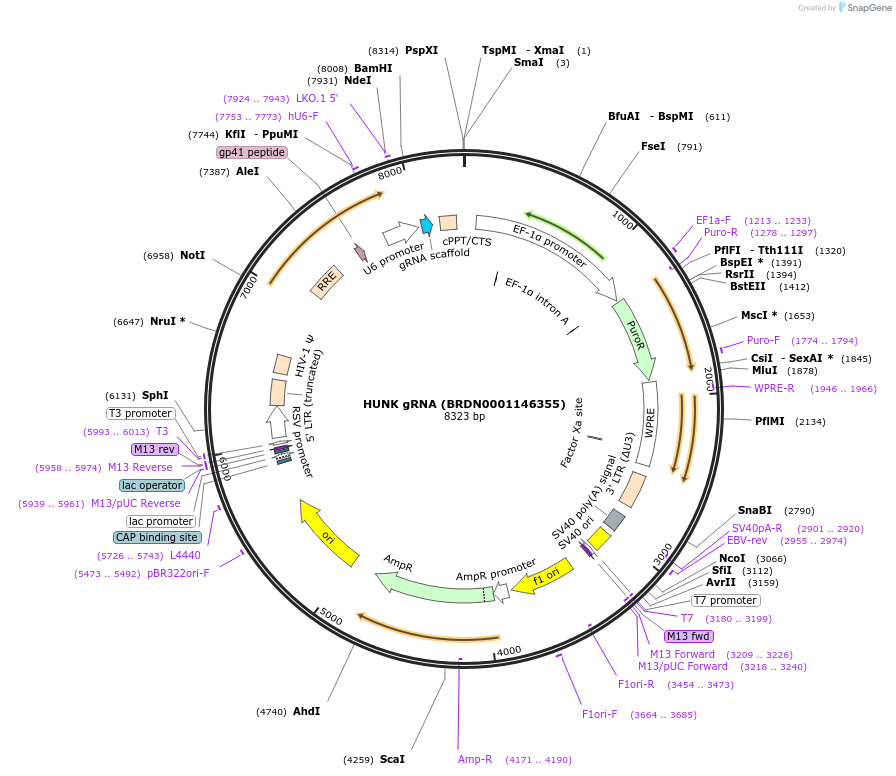
HUNK gRNA (BRDN0001146355)
Plasmid#75939Purpose3rd generation lentiviral gRNA plasmid targeting human HUNKDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
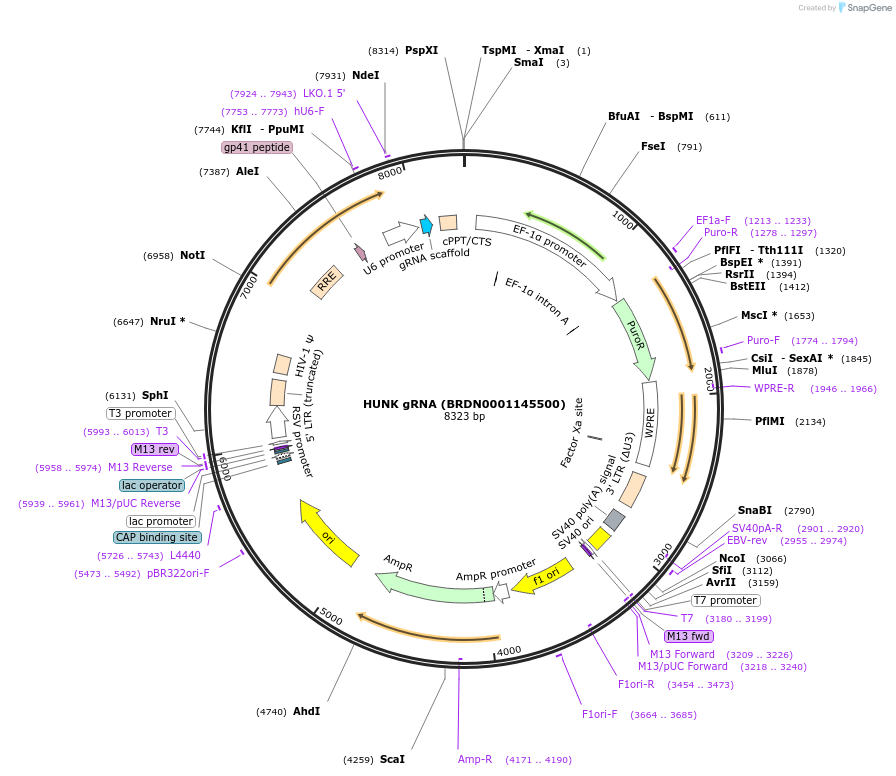
HUNK gRNA (BRDN0001145500)
Plasmid#75941Purpose3rd generation lentiviral gRNA plasmid targeting human HUNKDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
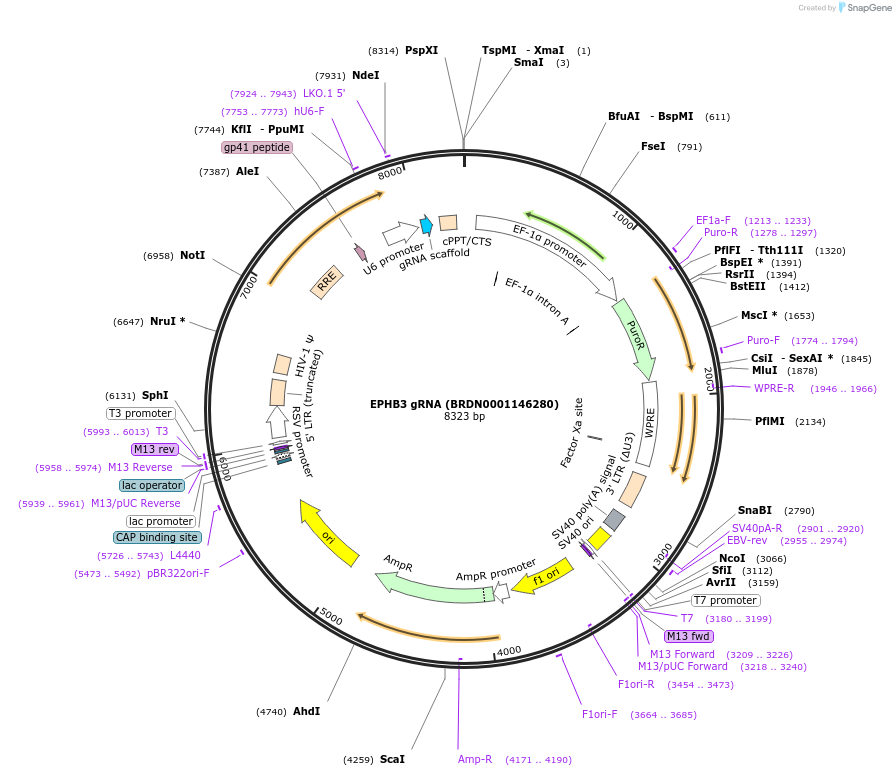
EPHB3 gRNA (BRDN0001146280)
Plasmid#75945Purpose3rd generation lentiviral gRNA plasmid targeting human EPHB3DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
EPHB3 gRNA (BRDN0001145322)
Plasmid#75946Purpose3rd generation lentiviral gRNA plasmid targeting human EPHB3DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
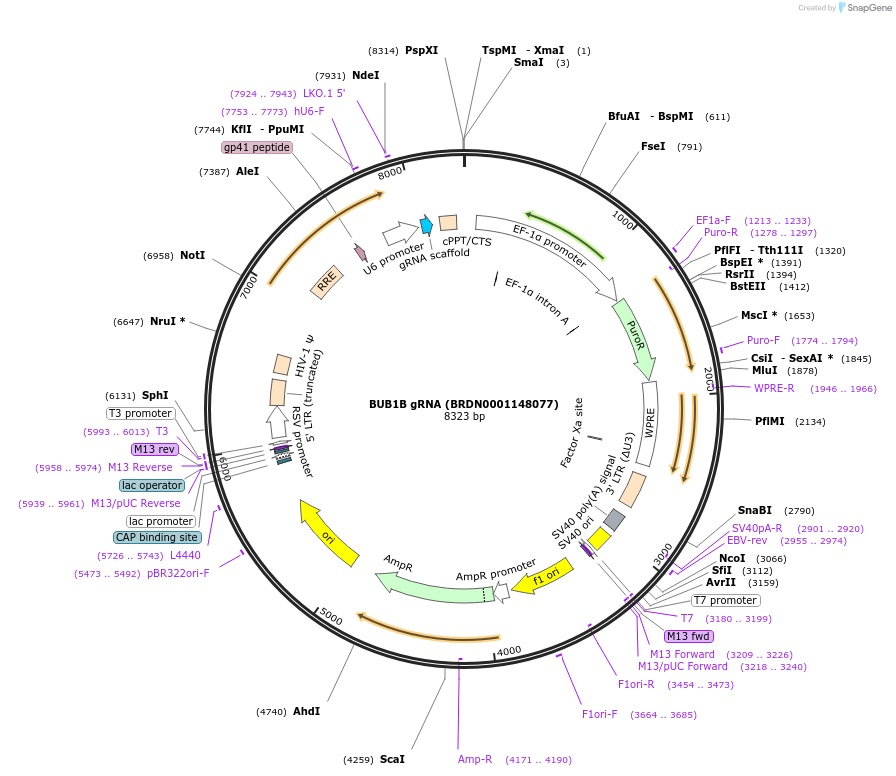
BUB1B gRNA (BRDN0001148077)
Plasmid#75918Purpose3rd generation lentiviral gRNA plasmid targeting human BUB1BDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
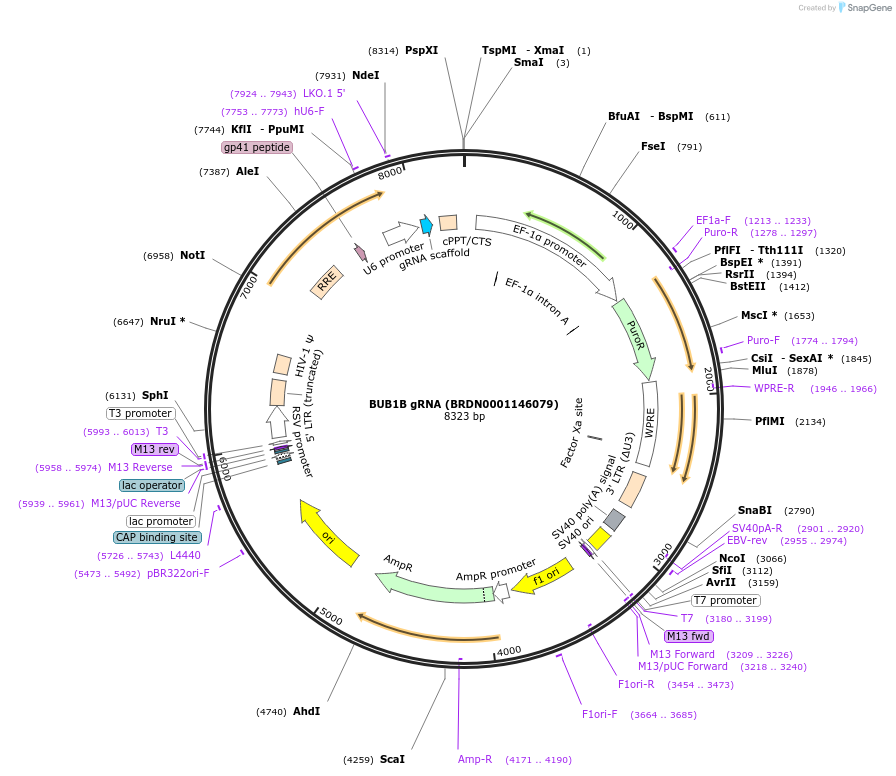
BUB1B gRNA (BRDN0001146079)
Plasmid#75919Purpose3rd generation lentiviral gRNA plasmid targeting human BUB1BDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
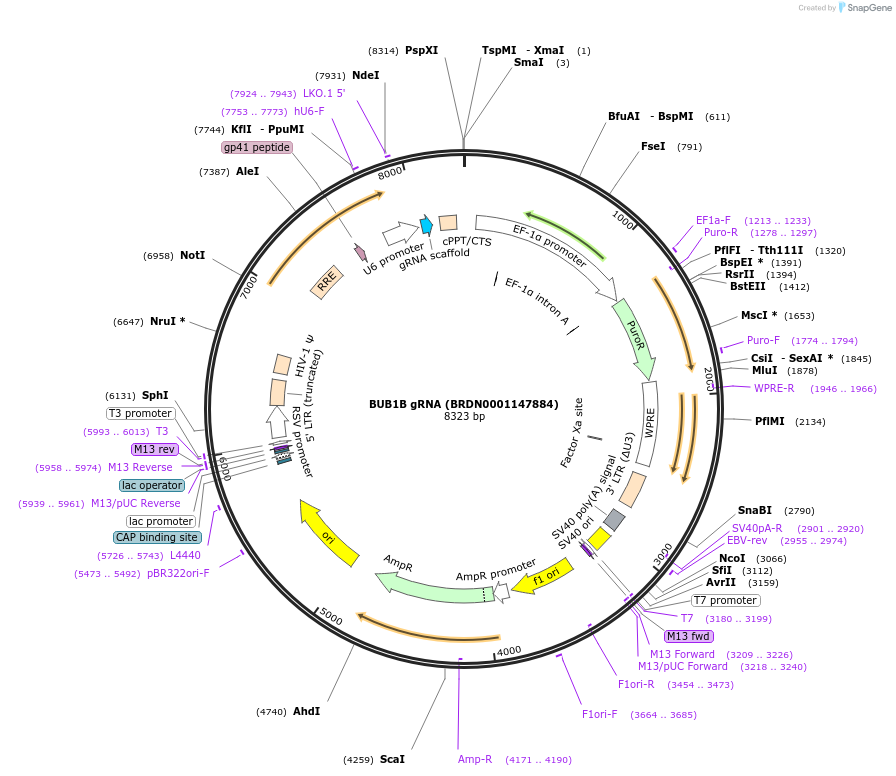
BUB1B gRNA (BRDN0001147884)
Plasmid#75920Purpose3rd generation lentiviral gRNA plasmid targeting human BUB1BDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
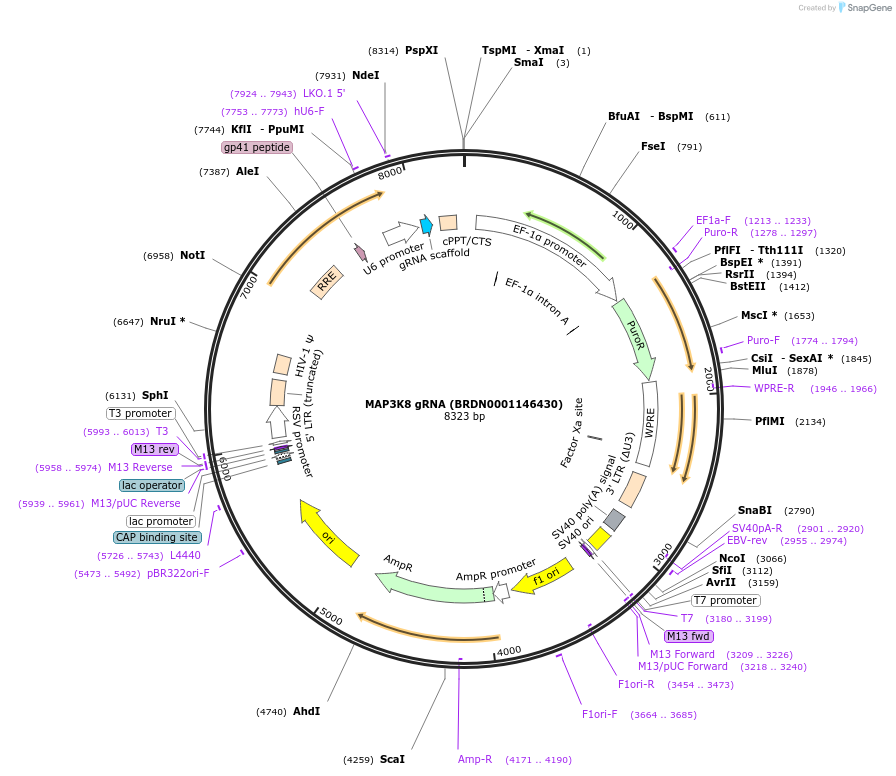
MAP3K8 gRNA (BRDN0001146430)
Plasmid#75921Purpose3rd generation lentiviral gRNA plasmid targeting human MAP3K8DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
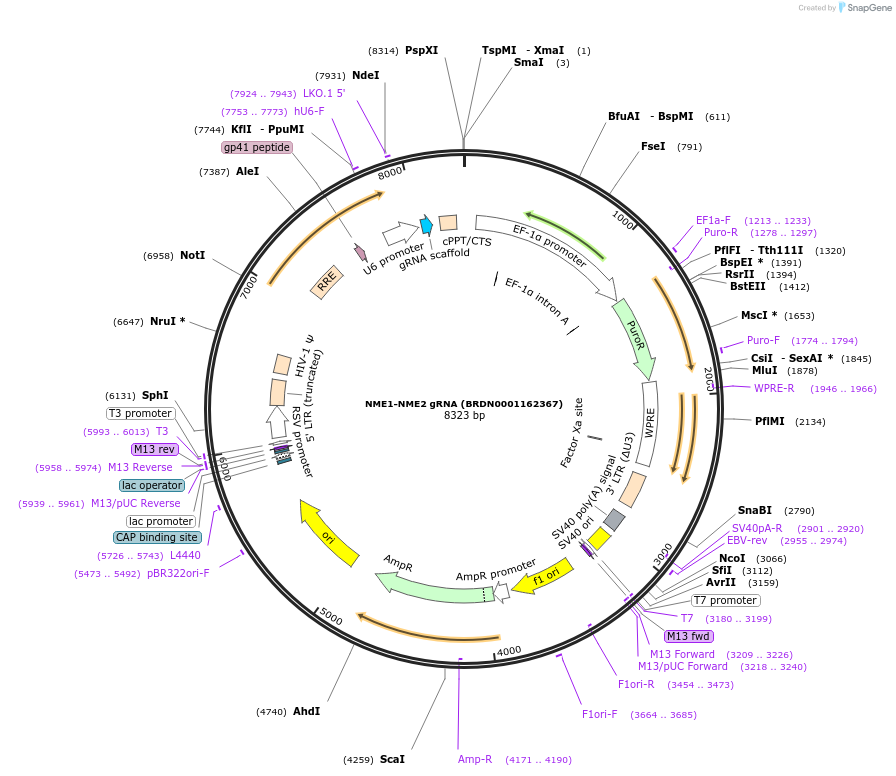
NME1-NME2 gRNA (BRDN0001162367)
Plasmid#75888Purpose3rd generation lentiviral gRNA plasmid targeting human NME1-NME2DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
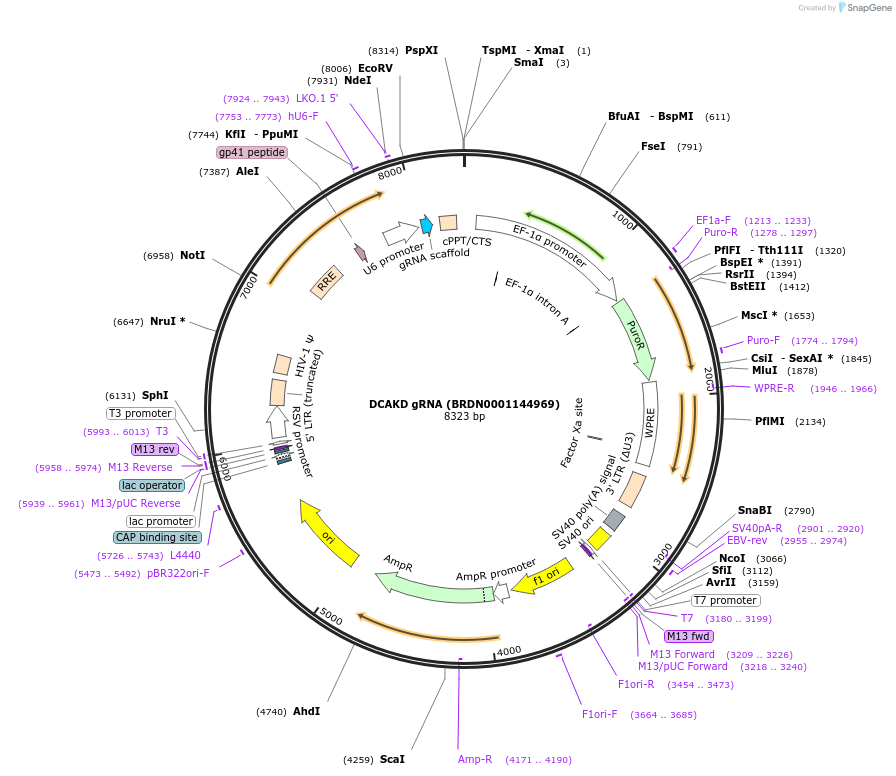
DCAKD gRNA (BRDN0001144969)
Plasmid#75872Purpose3rd generation lentiviral gRNA plasmid targeting human DCAKDDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
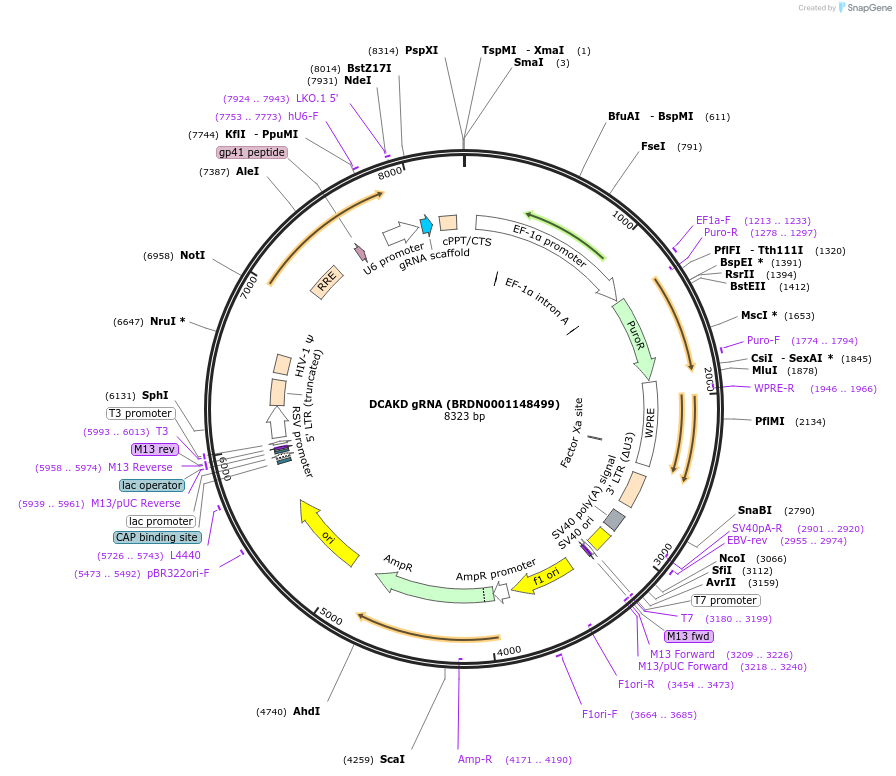
DCAKD gRNA (BRDN0001148499)
Plasmid#75873Purpose3rd generation lentiviral gRNA plasmid targeting human DCAKDDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
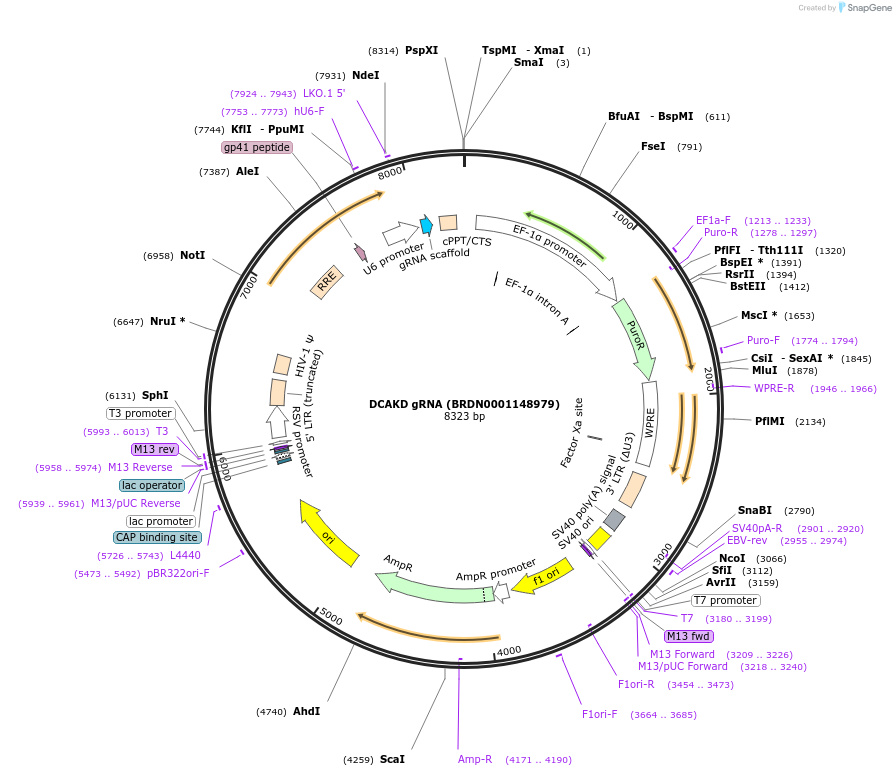
DCAKD gRNA (BRDN0001148979)
Plasmid#75874Purpose3rd generation lentiviral gRNA plasmid targeting human DCAKDDepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
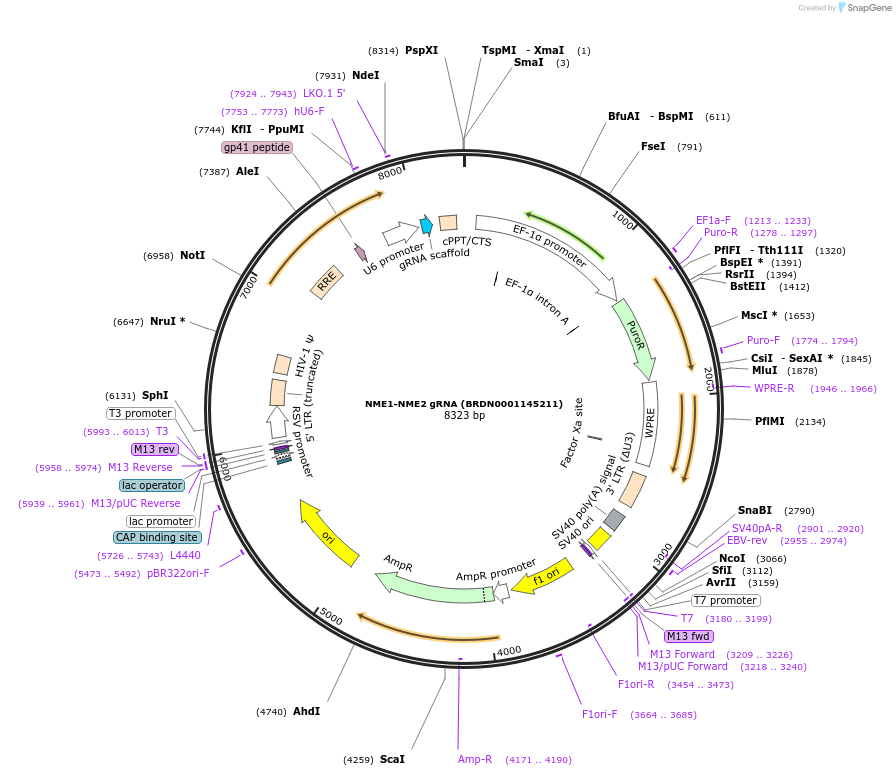
NME1-NME2 gRNA (BRDN0001145211)
Plasmid#75885Purpose3rd generation lentiviral gRNA plasmid targeting human NME1-NME2DepositorAvailable SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only