We narrowed to 17,800 results for: JUN
-
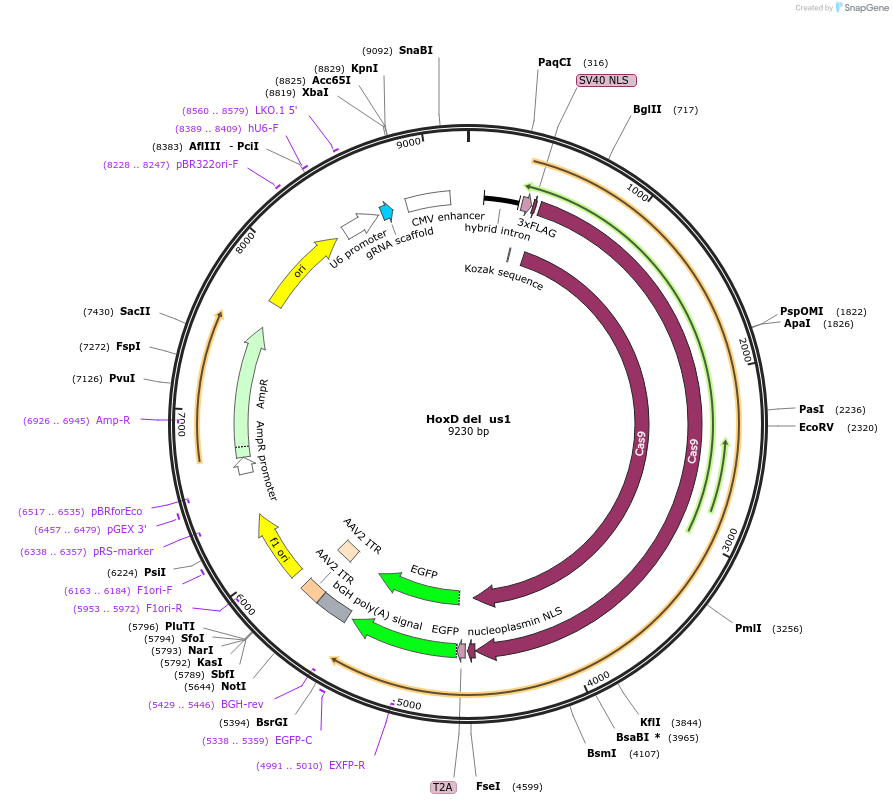
Plasmid#131337PurposegRNA to delete a nucleation site near HoxD. Use with HoxD del ds1DepositorInsertEVX2-gRNA-N-del
UseCRISPRExpressionMammalianAvailable SinceMarch 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
HoxD del control us
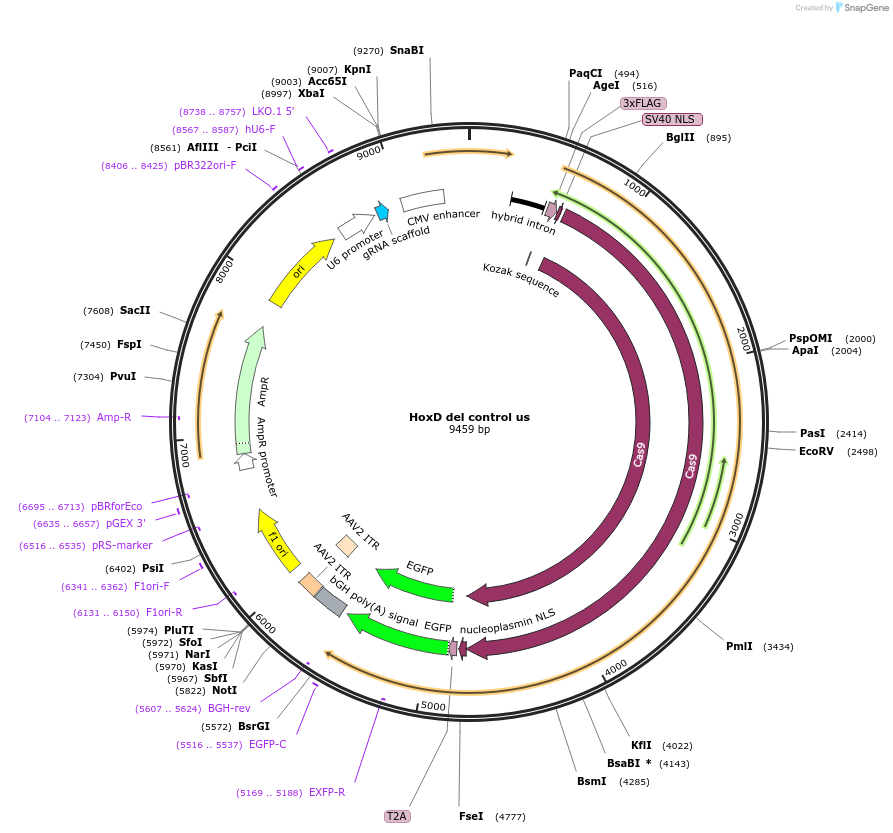
Plasmid#131335PurposegRNA to delete control region near HoxD. Use with HoxD del control dsDepositorInsertCTRL-gRNA-N-del
UseCRISPRExpressionMammalianAvailable SinceMarch 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
EED-KO-gRNA-1
Plasmid#131321PurposegRNA to knockout EED. Use with EED-KO-gRNA-2DepositorInsertEED-KO-gRNA-1
UseCRISPRExpressionMammalianAvailable SinceMarch 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
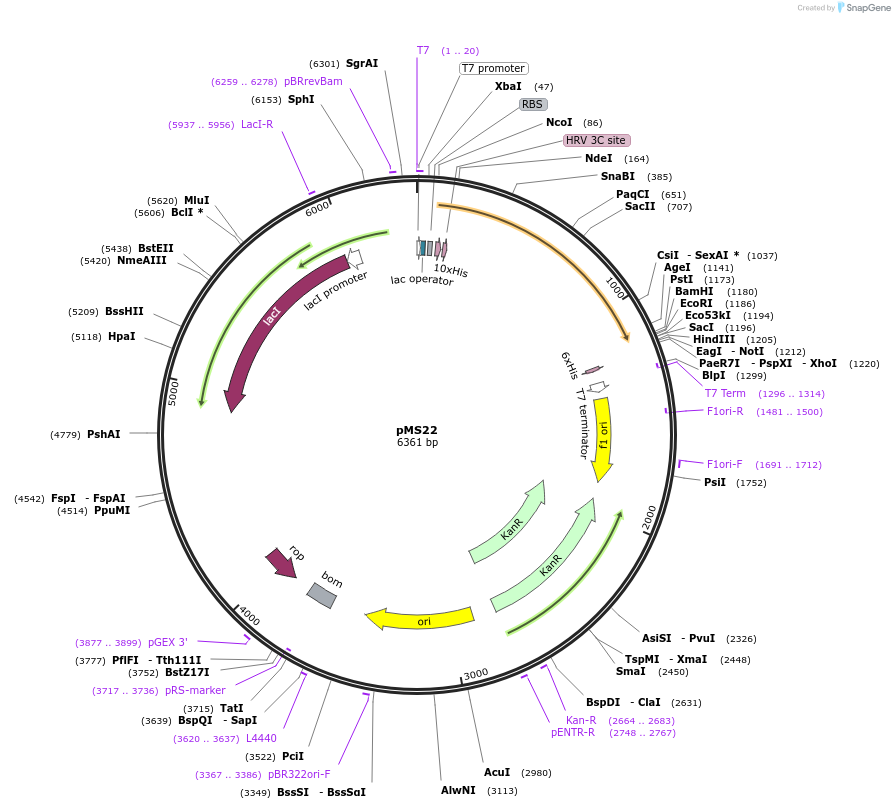
pMS22
Plasmid#129710PurposeExpresses Escherichia coli RssB E134G:E135S:E136G:E137S variant as an N-terminal 10His fusionDepositorInsertEscherichia coli RssB-GSGS variant (rssB Escherichia coli MG1655)
Tags10-Histidine tag cleavable with PreScission Prote…ExpressionBacterialMutationRssB E134G:E135S:E136G:E137SPromoterT7Available SinceAug. 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
pNH-TrxT-S18
Plasmid#127826PurposeProduction of recombinant human ribosomal protein S18DepositorAvailable SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
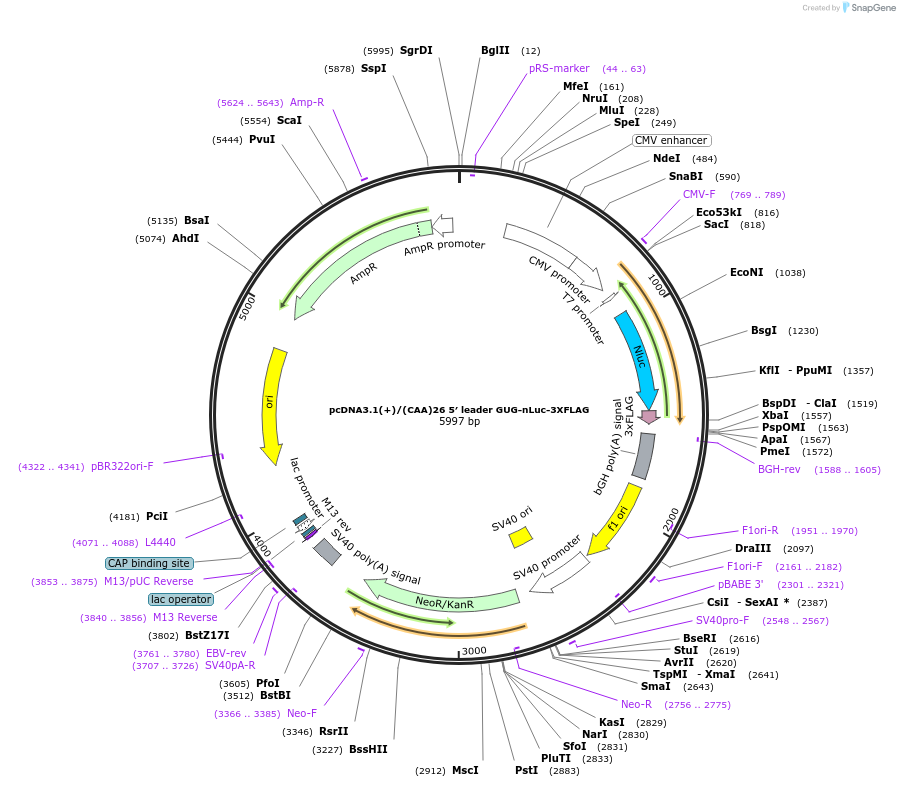
pcDNA3.1(+)/(CAA)26 5′ leader GUG-nLuc-3XFLAG
Plasmid#127325PurposeExpress 3xFLAG tagged nanoLuciferase (nLuc) from GUG start codon from mRNA with (CAA)26 repeats in 5' UTRDepositorInsertnanoLuciferase
Tags3xFLAGExpressionMammalianPromoterCMVAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
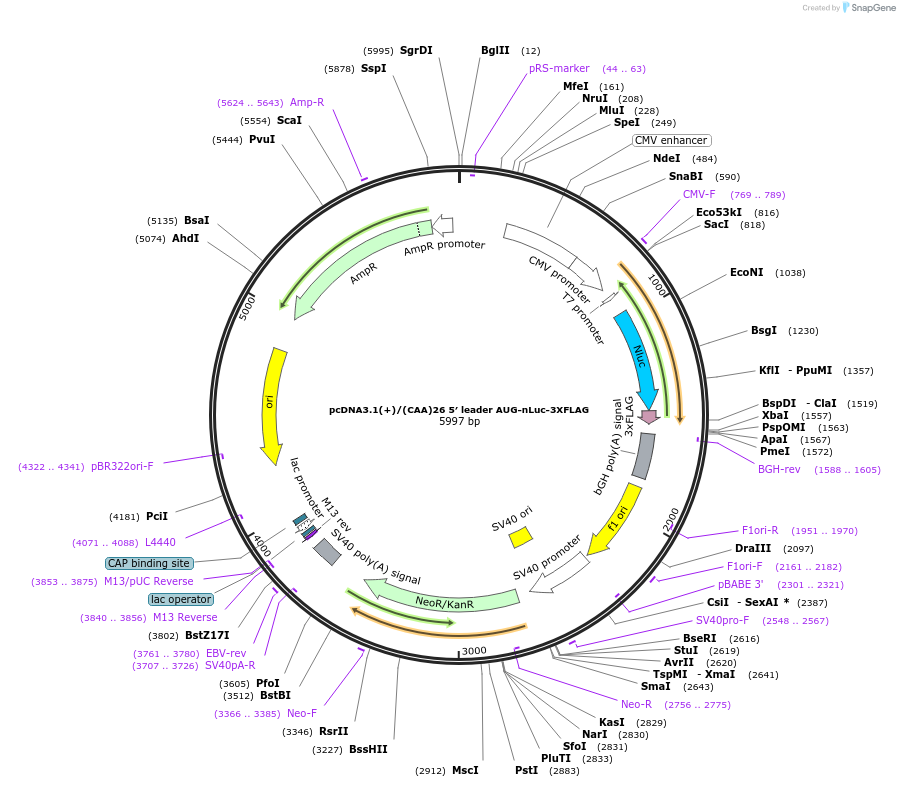
pcDNA3.1(+)/(CAA)26 5′ leader AUG-nLuc-3XFLAG
Plasmid#127324PurposeExpress 3xFLAG tagged nanoLuciferase (nLuc) from AUG start codon from mRNA with (CAA)26 repeats in 5' UTRDepositorInsertnanoLuciferase
Tags3xFLAGExpressionMammalianPromoterCMVAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
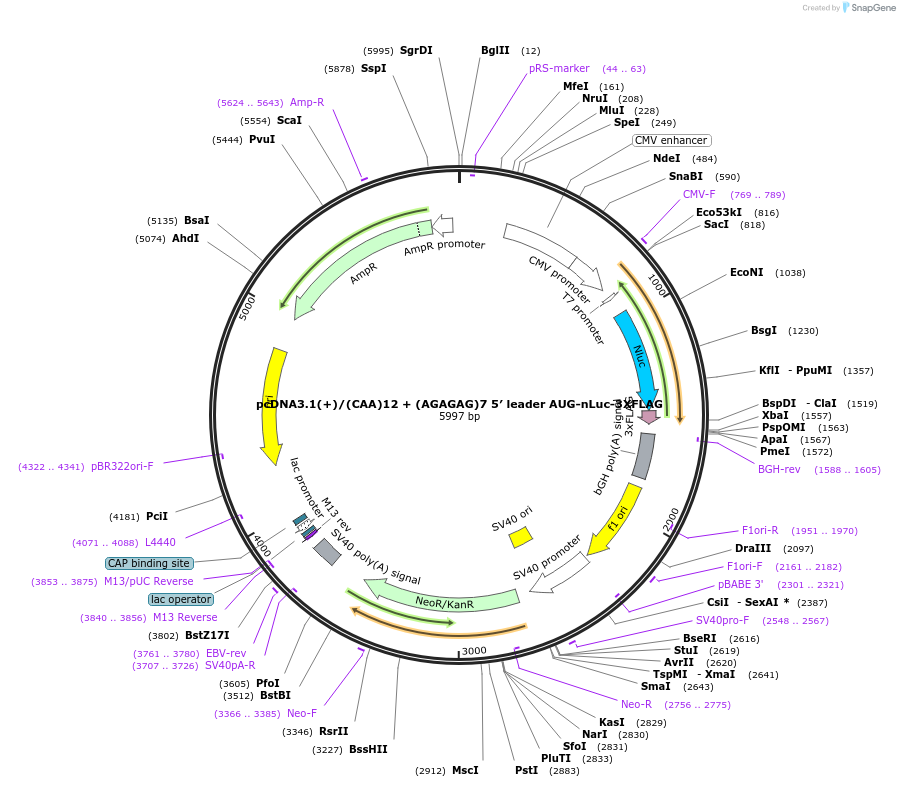
pcDNA3.1(+)/(CAA)12 + (AGAGAG)7 5′ leader GUG-nLuc-3XFLAG
Plasmid#127327PurposeExpress 3xFLAG tagged nanoLuciferase (nLuc) from GUG start codon from mRNA with (AGAGAG)7 repeats in 5' UTRDepositorInsertnanoLuciferase
Tags3xFLAGExpressionMammalianPromoterCMVAvailable SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
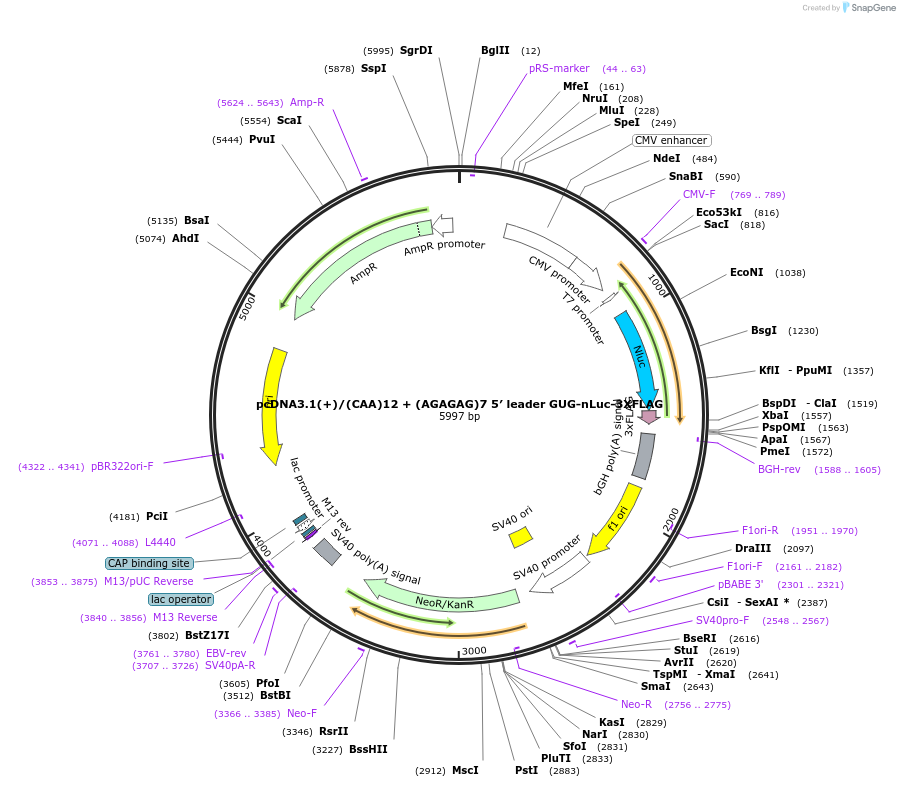
pcDNA3.1(+)/(CAA)12 + (AGAGAG)7 5′ leader AUG-nLuc-3XFLAG
Plasmid#127326PurposeExpress 3xFLAG tagged nanoLuciferase (nLuc) from AUG start codon from mRNA with (AGAGAG)7 repeats in 5' UTRDepositorInsertnanoLuciferase
Tags3xFLAGExpressionMammalianPromoterCMVAvailable SinceJune 24, 2019AvailabilityAcademic Institutions and Nonprofits only -
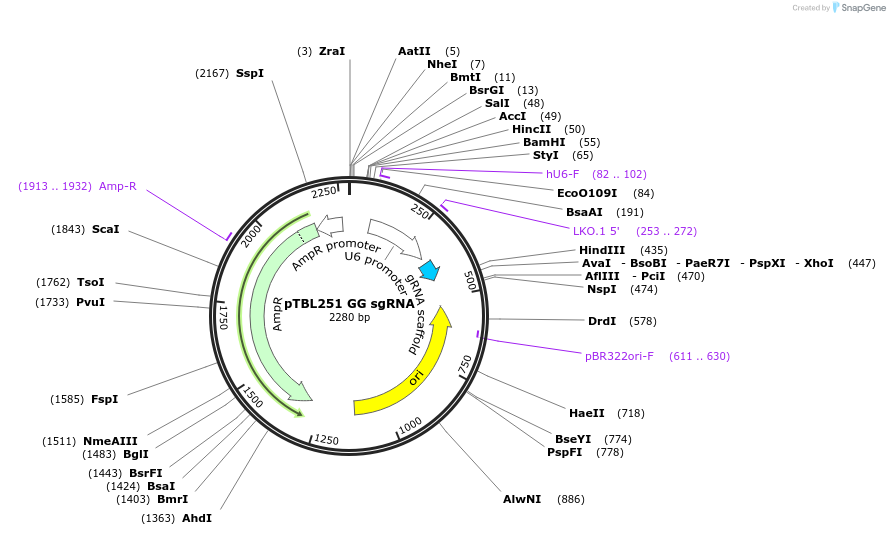
pTBL251 GG sgRNA
Plasmid#126436PurposeTo target a protospacer with GG at the cut site.DepositorInsertspacer against human genome with GG at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
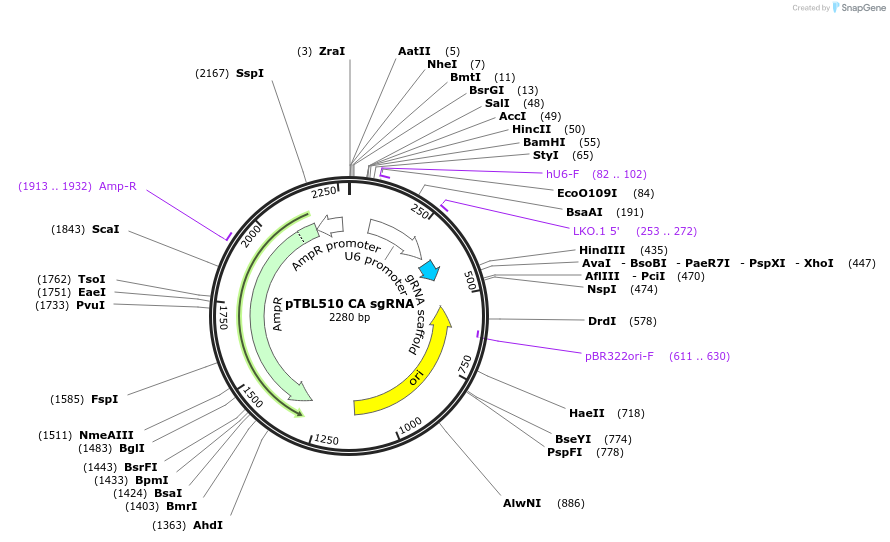
pTBL510 CA sgRNA
Plasmid#126430PurposeTo target a protospacer with CA at the cut site.DepositorInsertspacer against human genome with CA at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
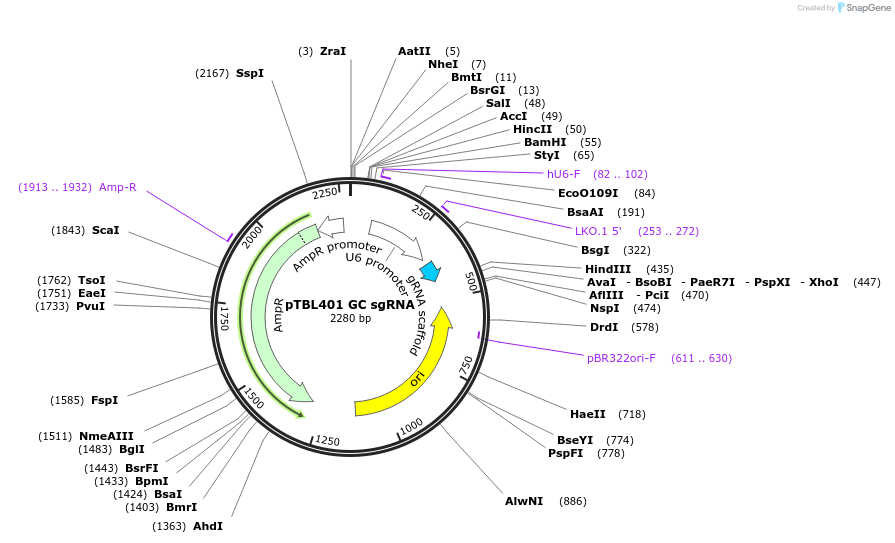
pTBL401 GC sgRNA
Plasmid#126435PurposeTo target a protospacer with GC at the cut site.DepositorInsertspacer against human genome with GC at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
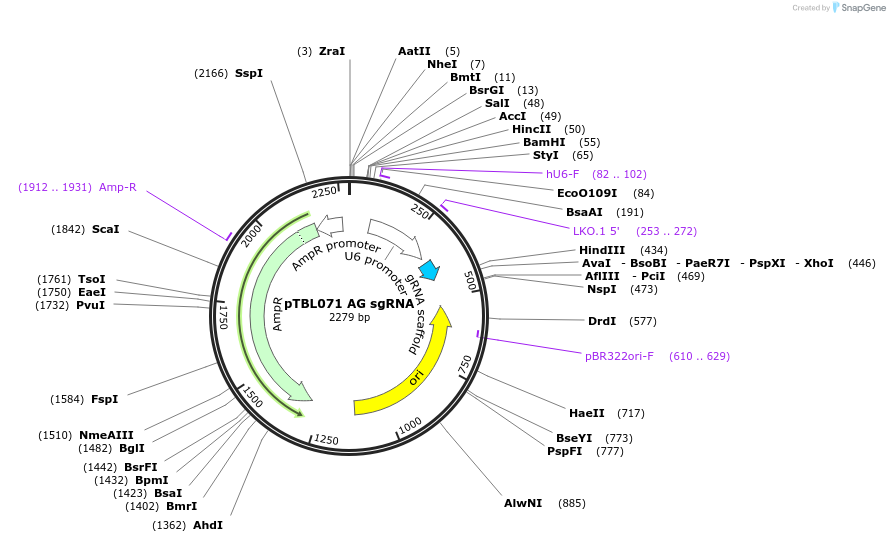
pTBL071 AG sgRNA
Plasmid#126428PurposeTo target a protospacer with AG at the cut site.DepositorInsertspacer against human genome with AG at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
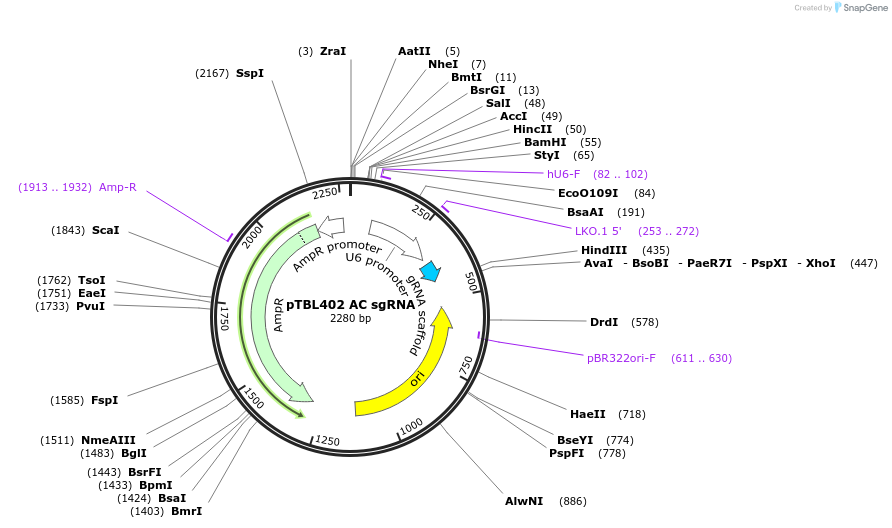
pTBL402 AC sgRNA
Plasmid#126427PurposeTo target a protospacer with AC at the cut site.DepositorInsertspacer against human genome with AC at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
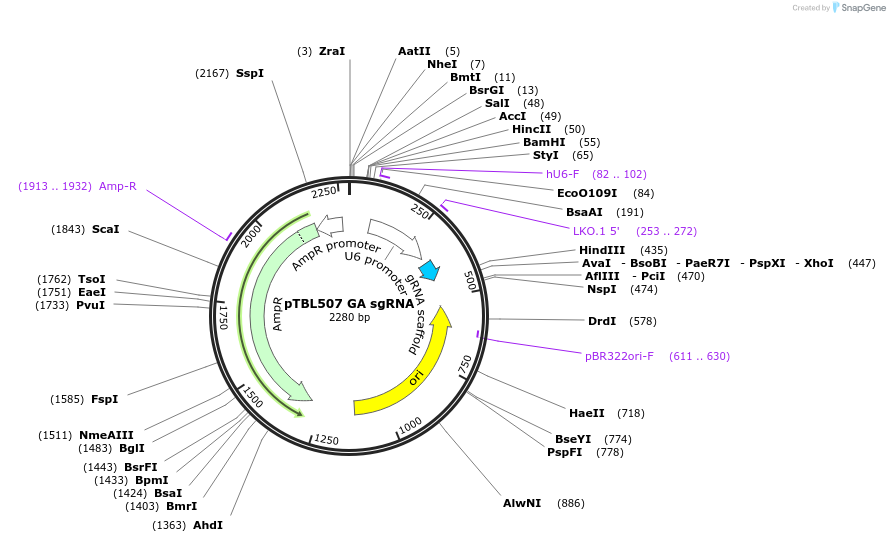
pTBL507 GA sgRNA
Plasmid#126434PurposeTo target a protospacer with GA at the cut site.DepositorInsertspacer against human genome with GA at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
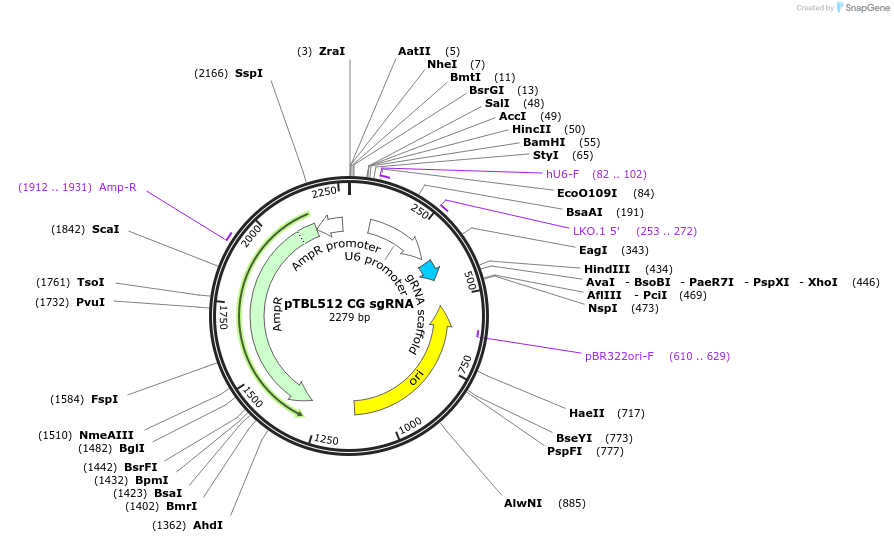
pTBL512 CG sgRNA
Plasmid#126432PurposeTo target a protospacer with CG at the cut site.DepositorInsertspacer against human genome with CG at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
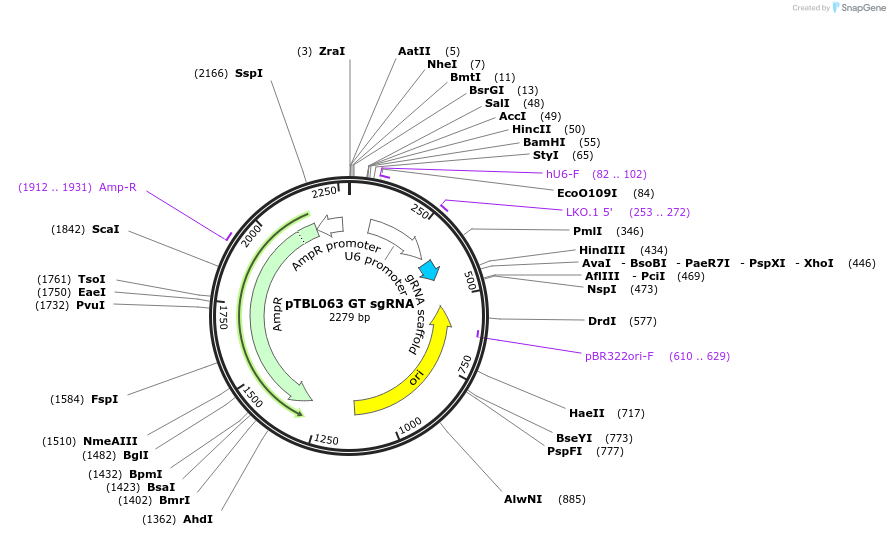
pTBL063 GT sgRNA
Plasmid#126437PurposeTo target a protospacer with GT at the cut site.DepositorInsertspacer against human genome with GT at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
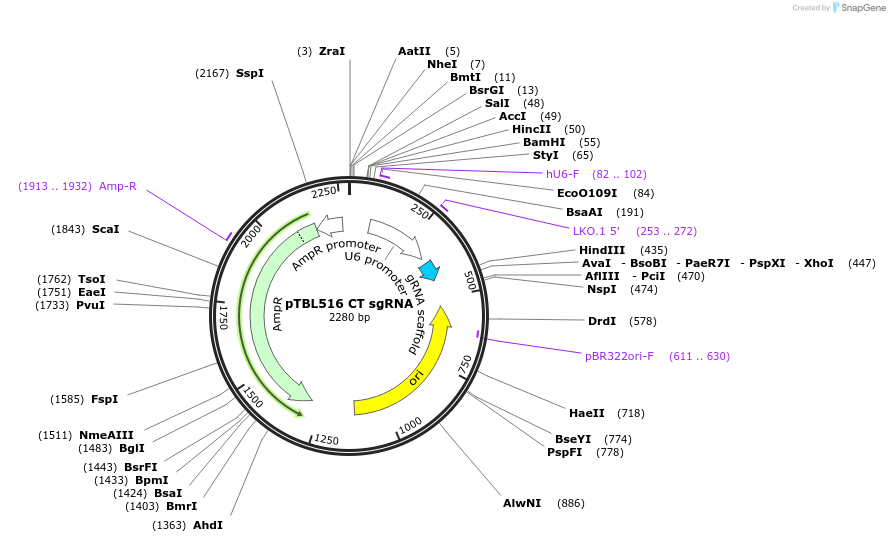
pTBL516 CT sgRNA
Plasmid#126433PurposeTo target a protospacer with CT at the cut site.DepositorInsertspacer against human genome with CT at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
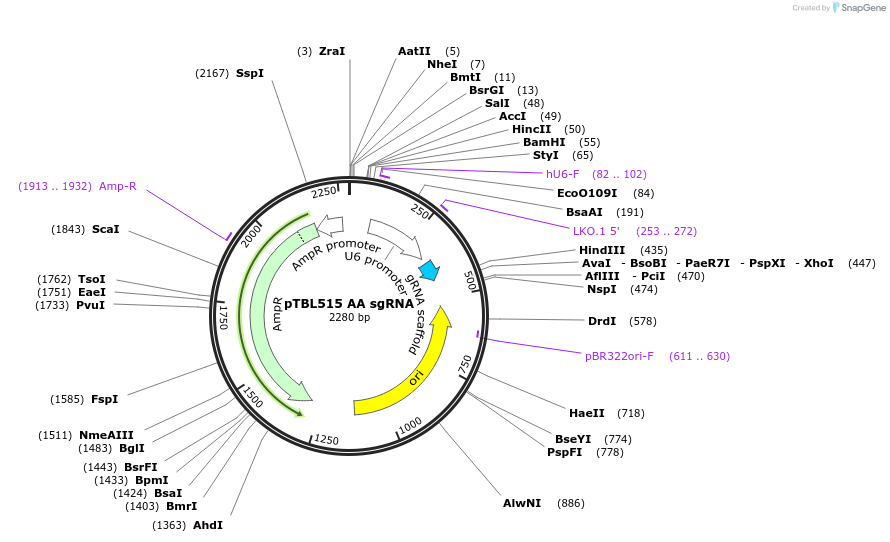
pTBL515 AA sgRNA
Plasmid#126426PurposeTo target a protospacer with AA at the cut site.DepositorInsertspacer against human genome with AA at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
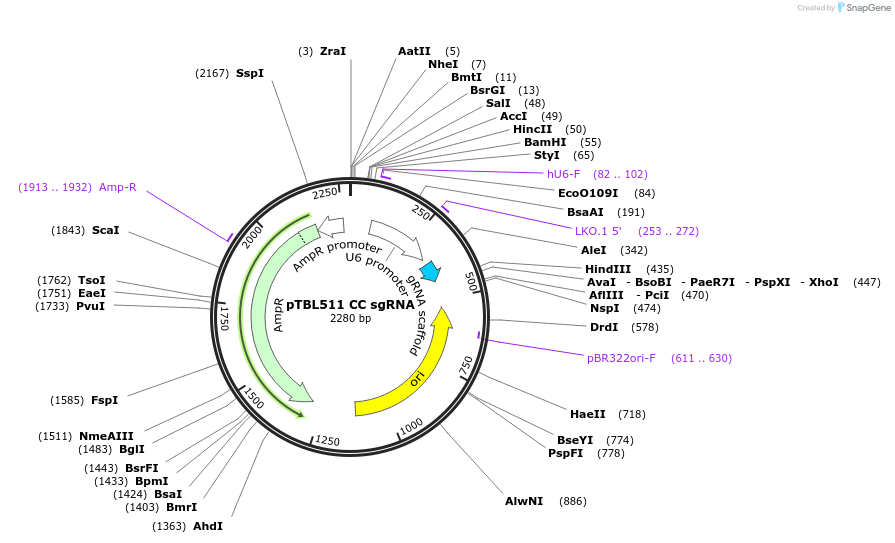
pTBL511 CC sgRNA
Plasmid#126431PurposeTo target a protospacer with CC at the cut site.DepositorInsertspacer against human genome with CC at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
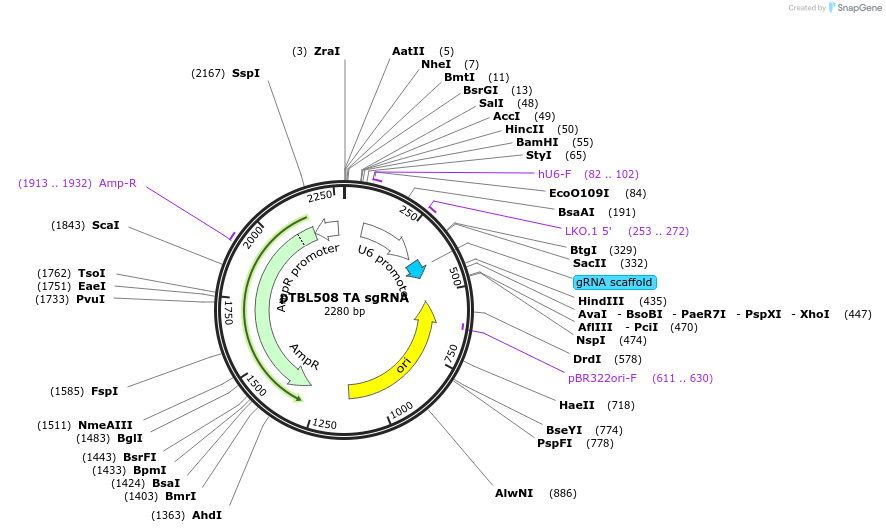
pTBL508 TA sgRNA
Plasmid#126438PurposeTo target a protospacer with TA at the cut site.DepositorInsertspacer against human genome with TA at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
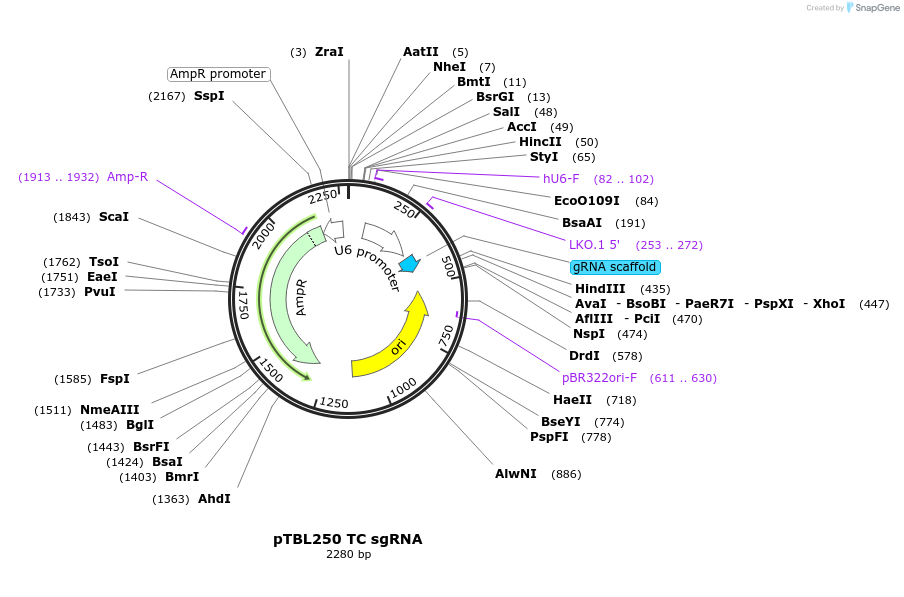
pTBL250 TC sgRNA
Plasmid#126439PurposeTo target a protospacer with TC at the cut site.DepositorInsertspacer against human genome with TC at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
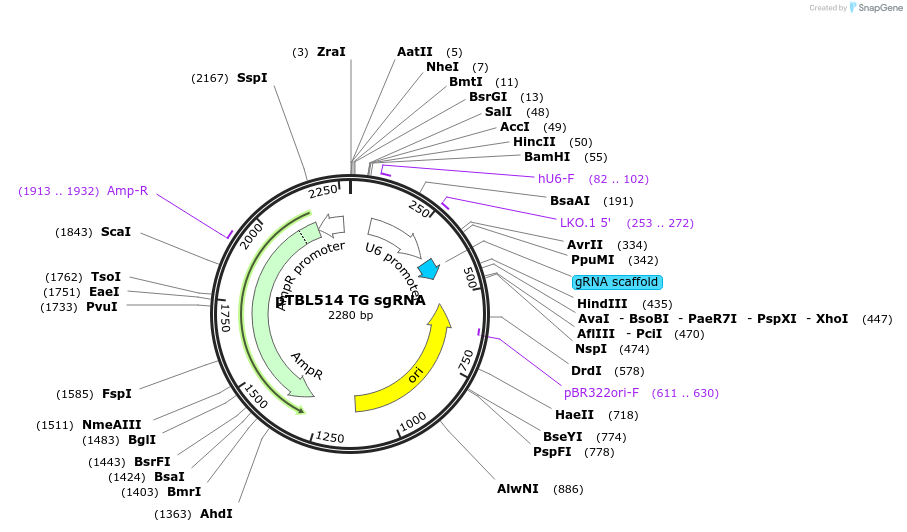
pTBL514 TG sgRNA
Plasmid#126440PurposeTo target a protospacer with TG at the cut site.DepositorInsertspacer against human genome with TG at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
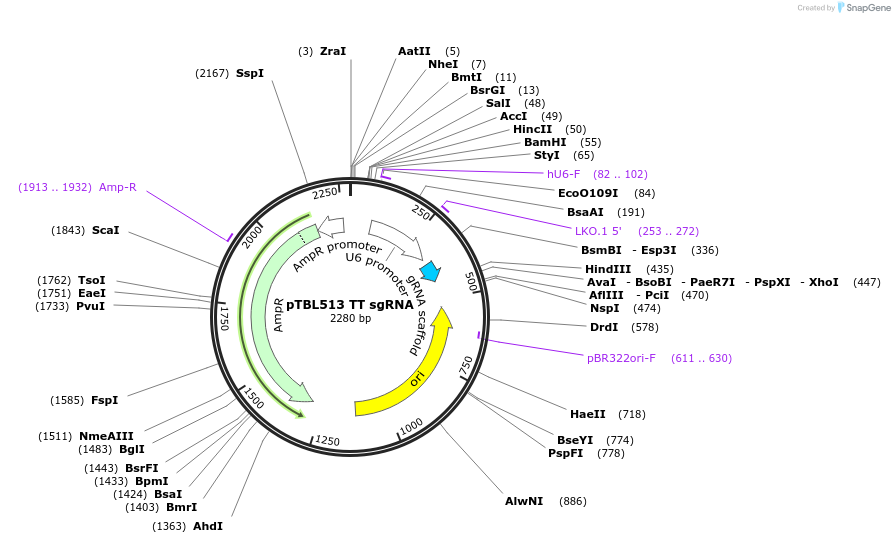
pTBL513 TT sgRNA
Plasmid#126441PurposeTo target a protospacer with TT at the cut site.DepositorInsertspacer against human genome with TT at cut site
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
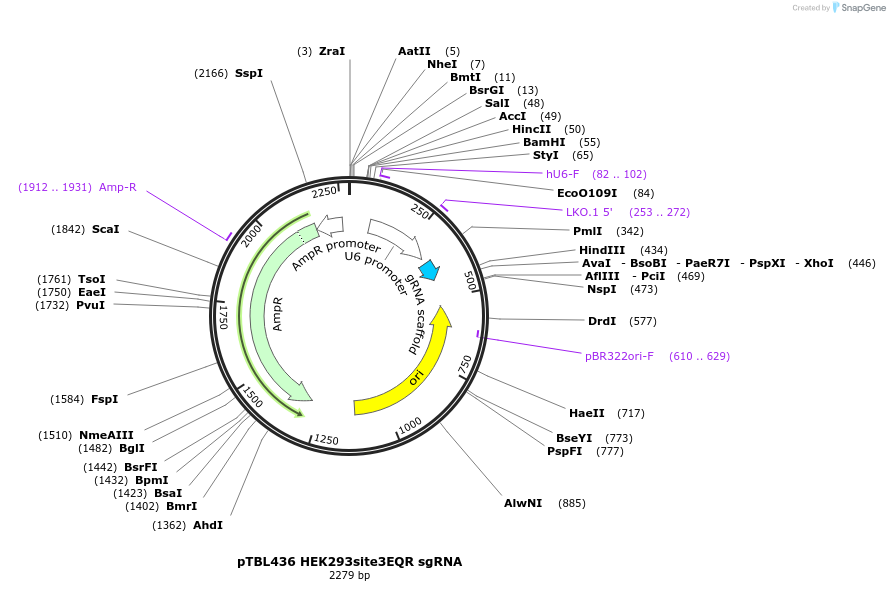
pTBL436 HEK293site3EQR sgRNA
Plasmid#126445PurposeTo cut the human HEK293 site3 locus for integration.DepositorInsertspacer against human HEK293 site3 with NGAG PAM
ExpressionMammalianPromoterhuman U6Available SinceJune 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
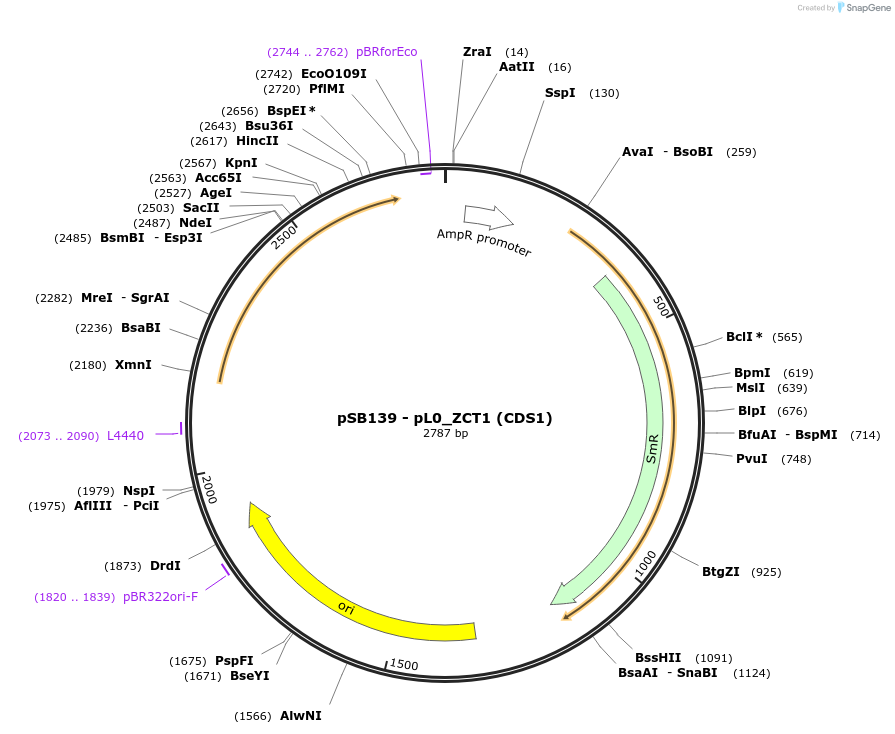
pSB139 - pL0_ZCT1 (CDS1)
Plasmid#123184PurposeGolden Gate (MoClo; CDS1) compatible ZCT1 gene from Catharanthus roseus; a repressor of the STR1 promoter from C. roseusDepositorInsertZCT1 from Catharanthus roseus
UsePart for plant expressionExpressionPlantMutationNo BpiI and BsaI sitesAvailable SinceMay 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
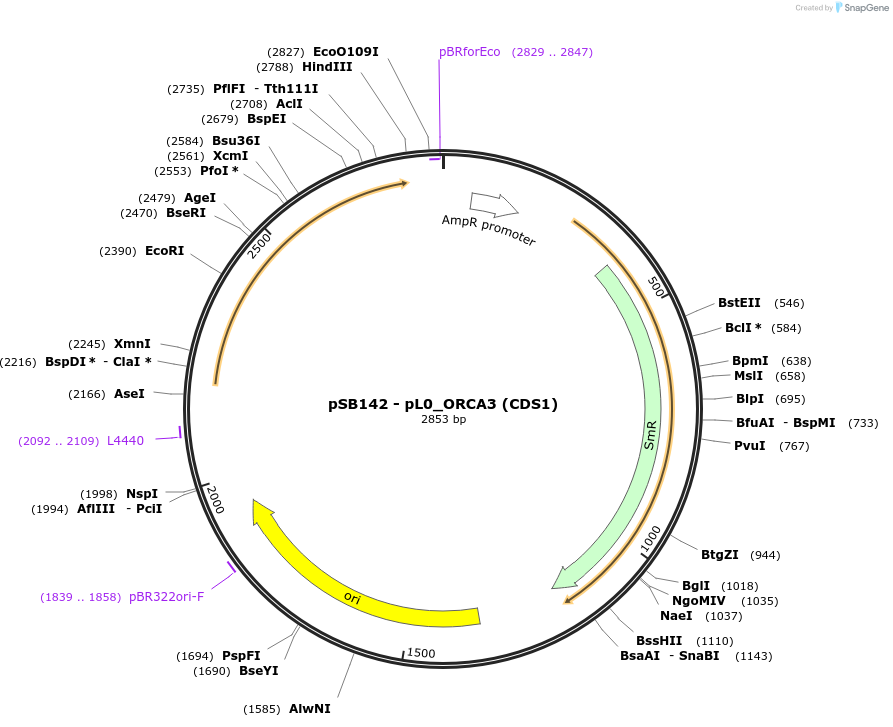
pSB142 - pL0_ORCA3 (CDS1)
Plasmid#123185PurposeGolden Gate (MoClo; CDS1) compatible ORCA3 gene from Catharanthus roseus; an activator of the STR1 promoter from C. roseusDepositorInsertORCA3 from Catharanthus roseus
UsePart for plant expressionExpressionPlantMutationNo BpiI and BsaI sitesAvailable SinceMay 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
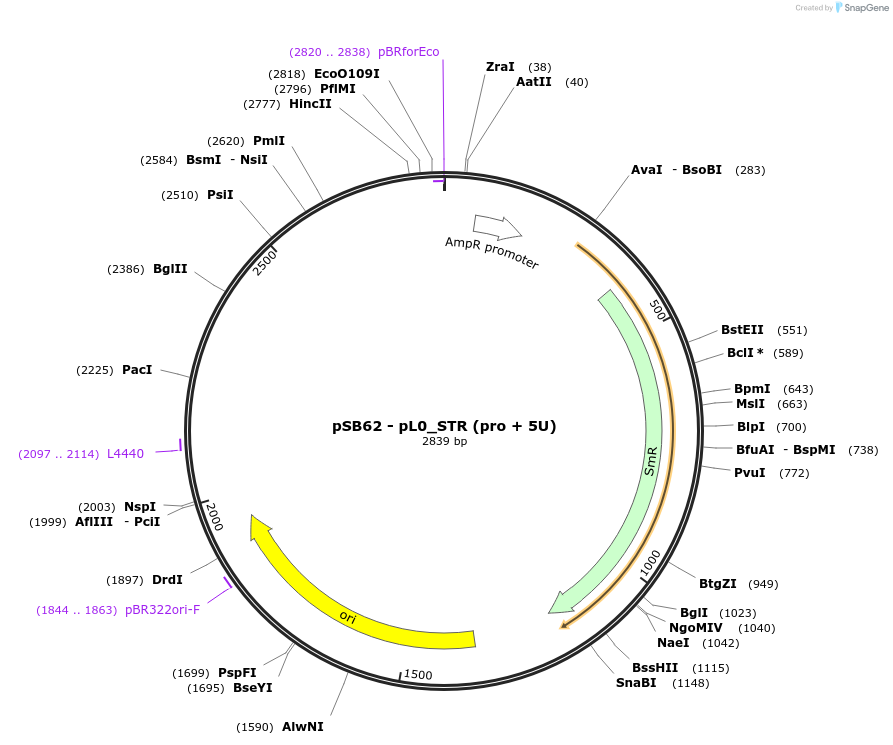
pSB62 - pL0_STR (pro + 5U)
Plasmid#123186PurposeGolden Gate (MoClo; PRO + 5U) compatible STR1 promoter from Catharanthus roseusDepositorInsertCatharanthus roseus STR1 promoter and 5'UTR
UsePart for plant expressionExpressionPlantMutationNo BpiI and BsaI sitesAvailable SinceMay 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
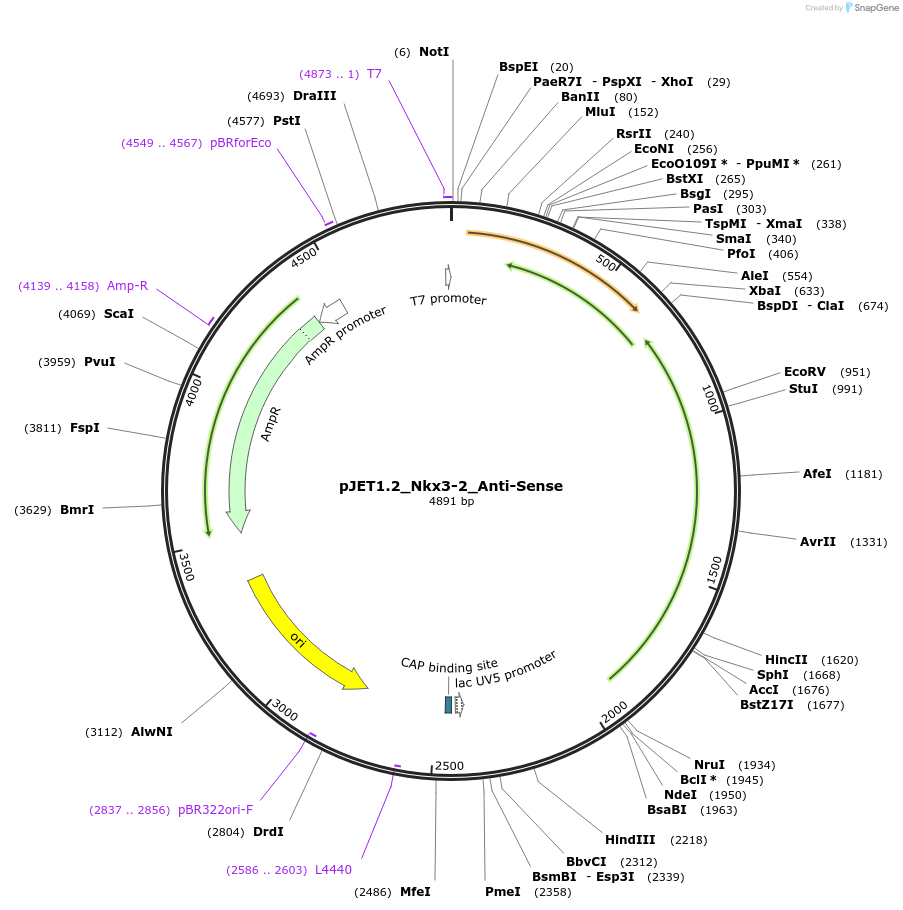
pJET1.2_Nkx3-2_Anti-Sense
Plasmid#124438PurposePlasmid for anti-sense in situ probe in vitro transcriptionDepositorAvailable SinceMay 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
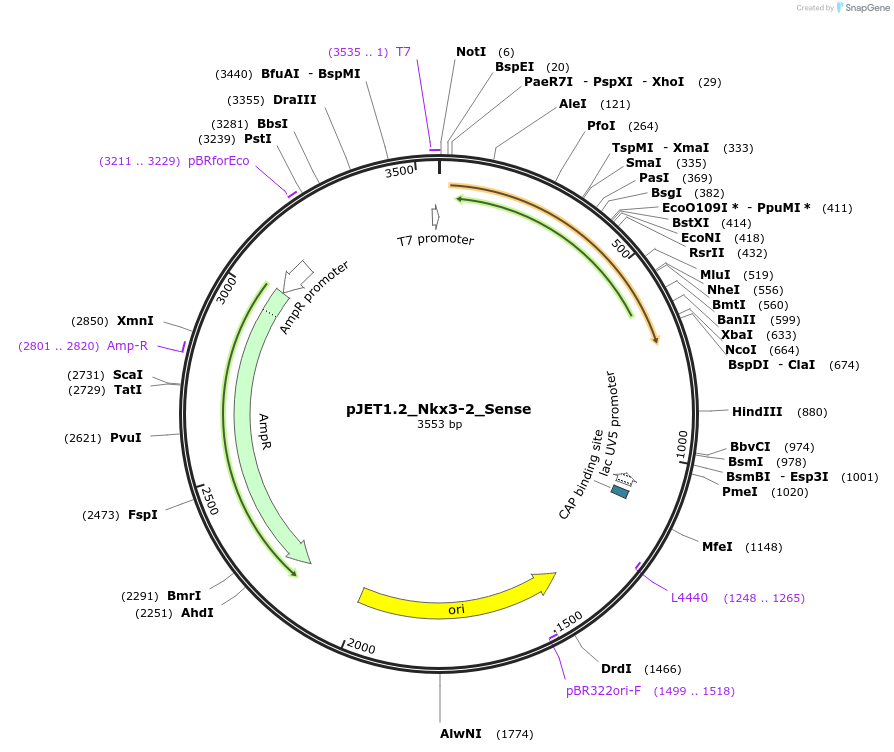
pJET1.2_Nkx3-2_Sense
Plasmid#124437PurposePlasmid for sense in situ probe in vitro transcriptionDepositorAvailable SinceMay 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
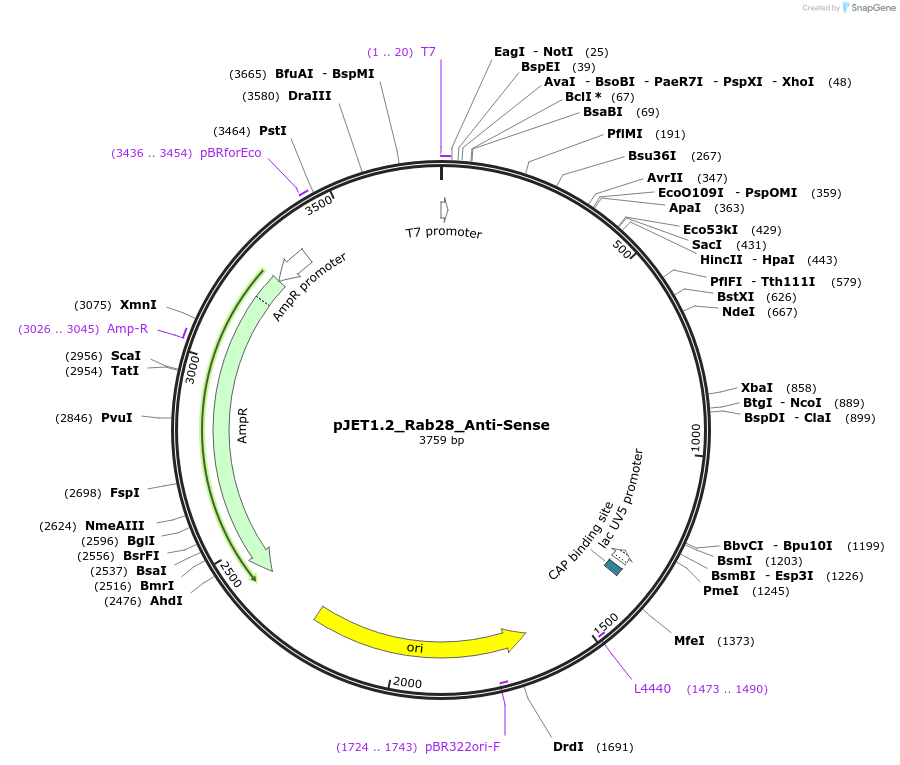
pJET1.2_Rab28_Anti-Sense
Plasmid#124442PurposePlasmid for anti-sense in situ probe in vitro transcriptionDepositorAvailable SinceMay 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
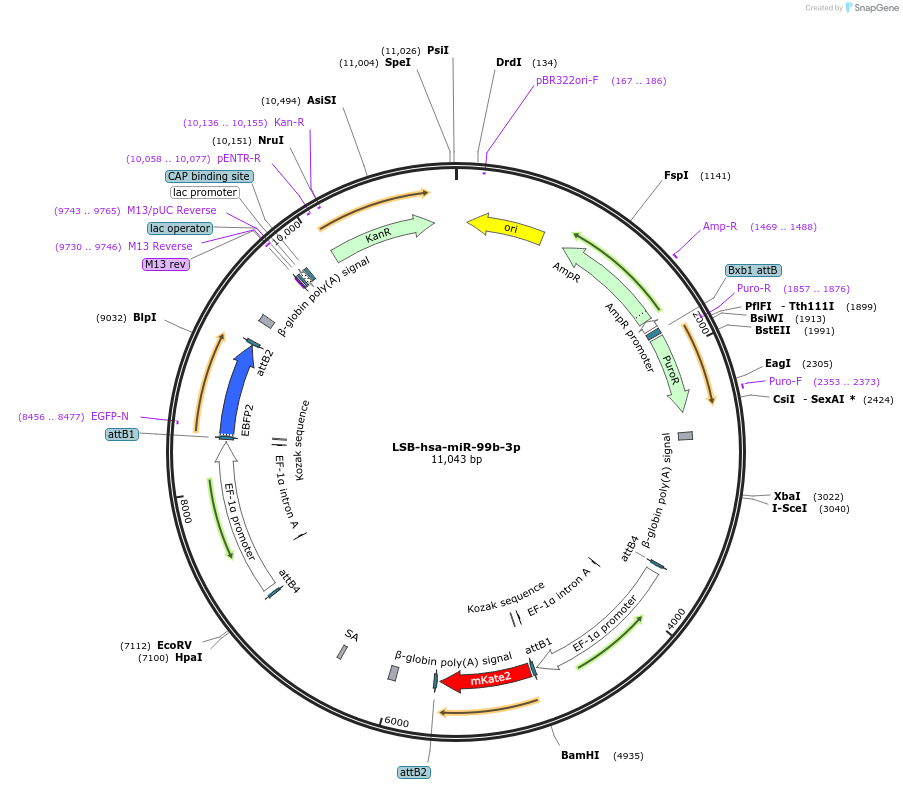
LSB-hsa-miR-99b-3p
Plasmid#103766PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-99b-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-99b-3p target (MIR99B Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
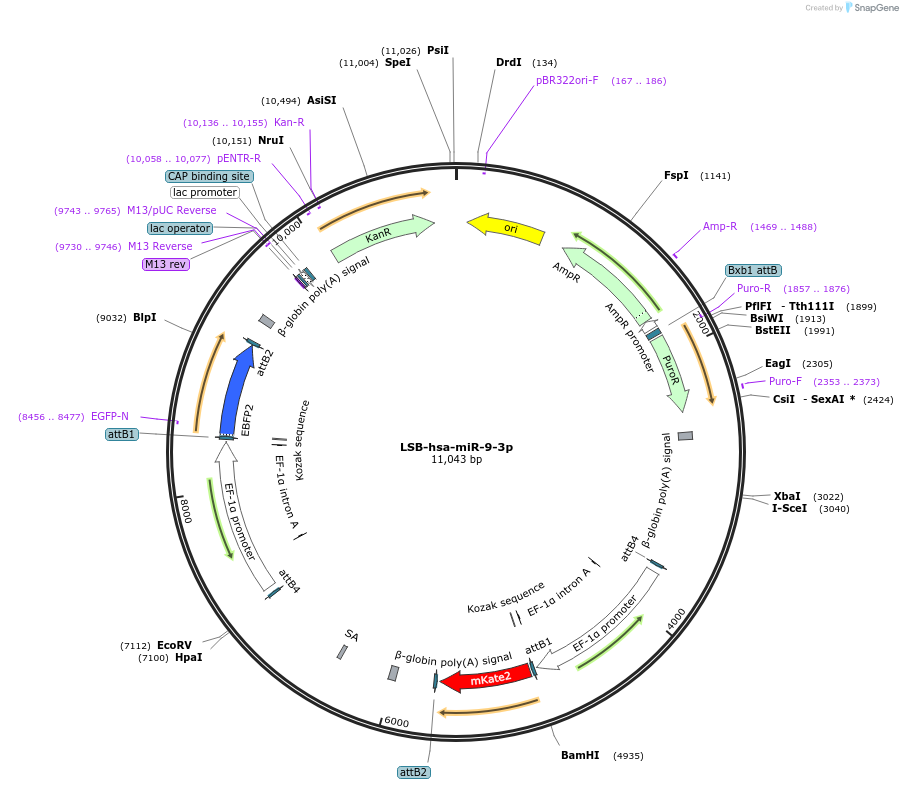
LSB-hsa-miR-9-3p
Plasmid#103748PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-9-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-9-3p target
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
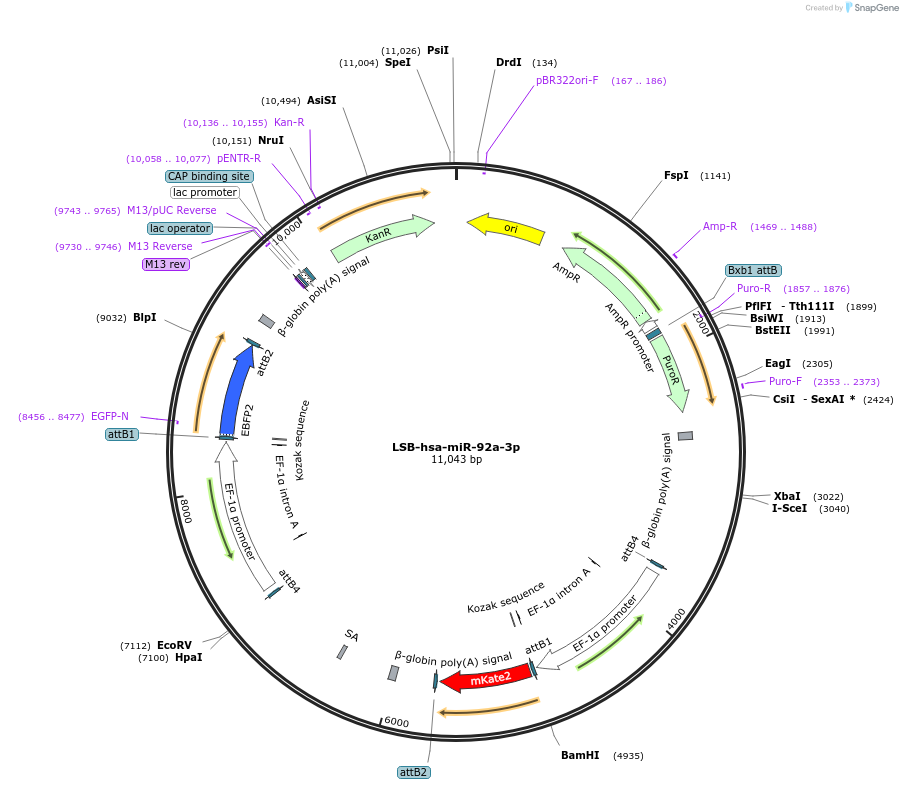
LSB-hsa-miR-92a-3p
Plasmid#103752PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-92a-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-92a-3p target
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
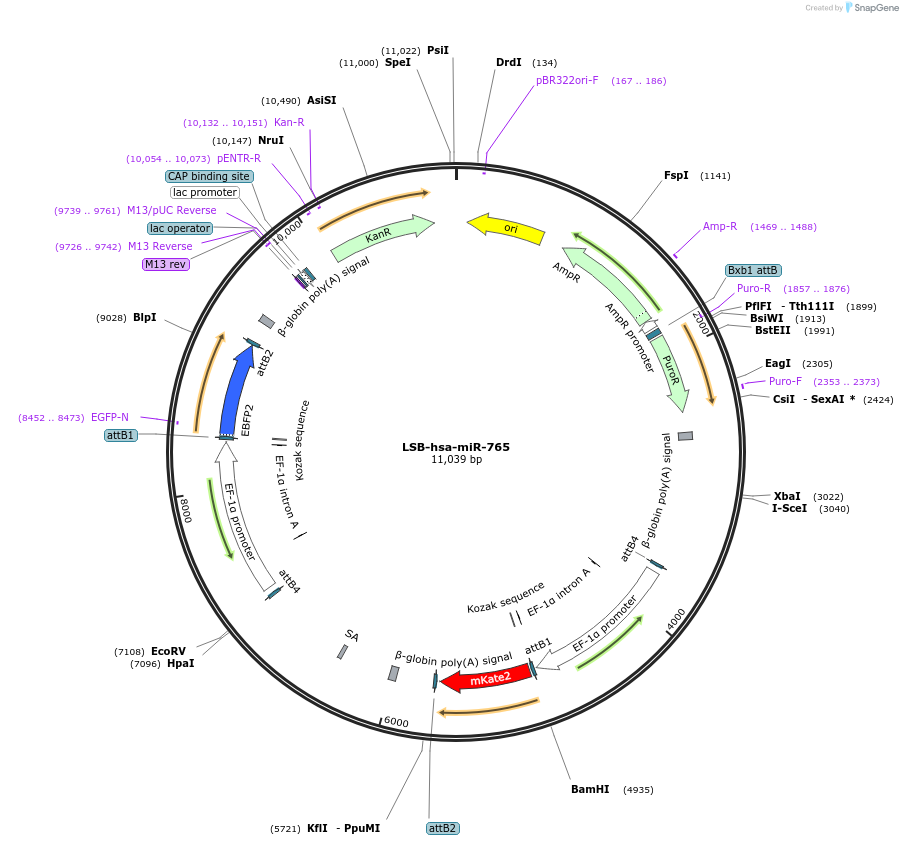
LSB-hsa-miR-765
Plasmid#103730PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-765 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
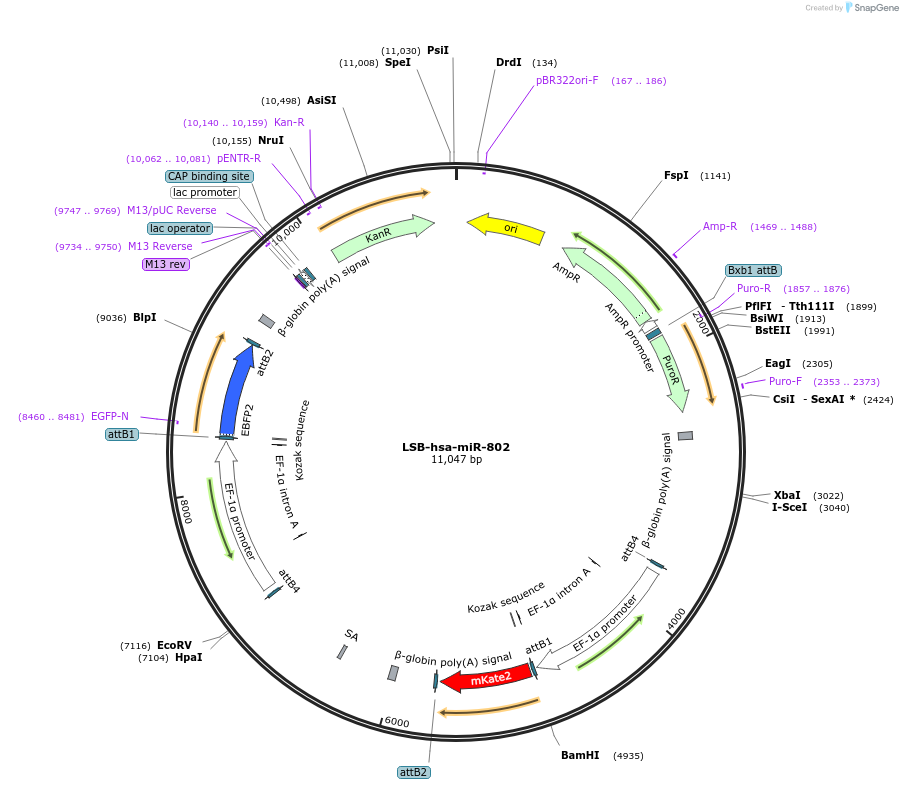
LSB-hsa-miR-802
Plasmid#103737PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-802 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
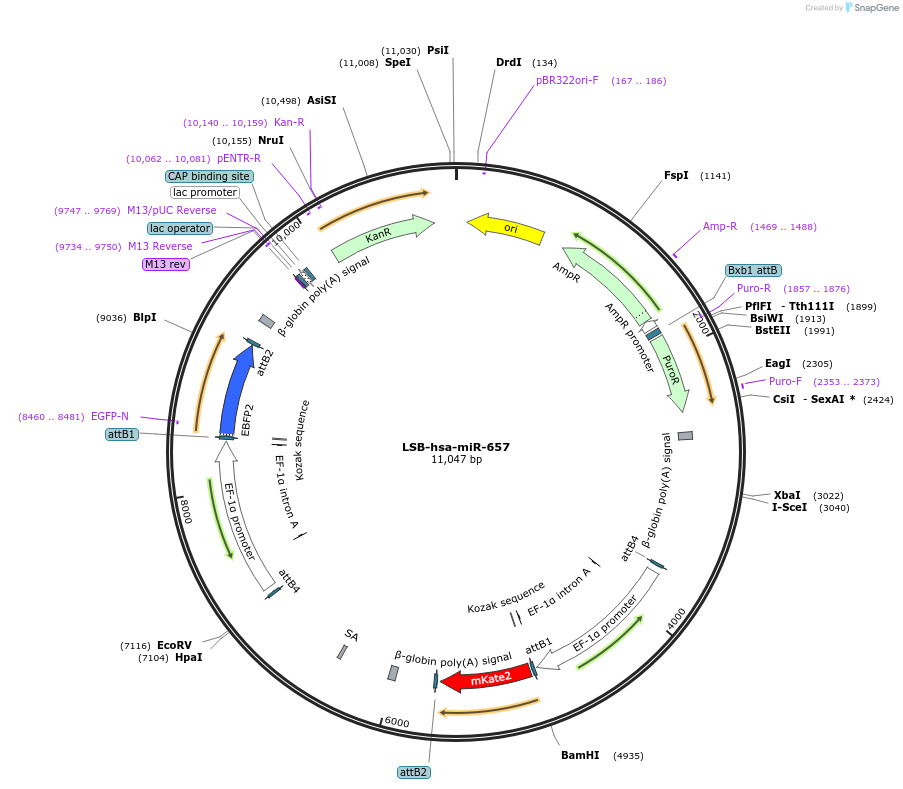
LSB-hsa-miR-657
Plasmid#103712PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-657 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
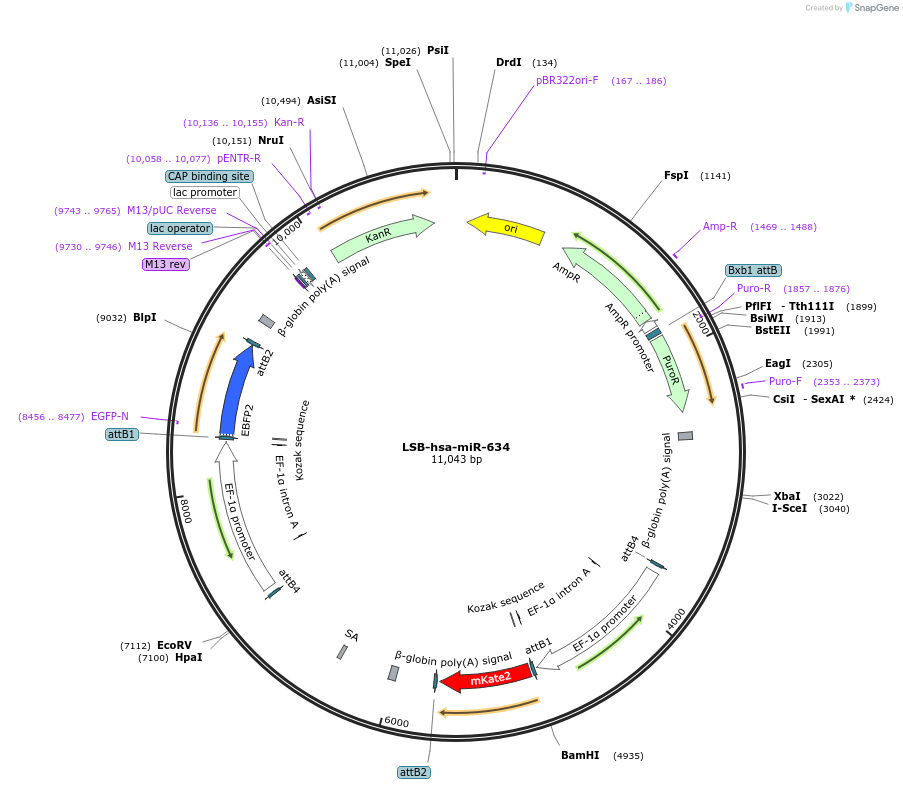
LSB-hsa-miR-634
Plasmid#103692PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-634 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
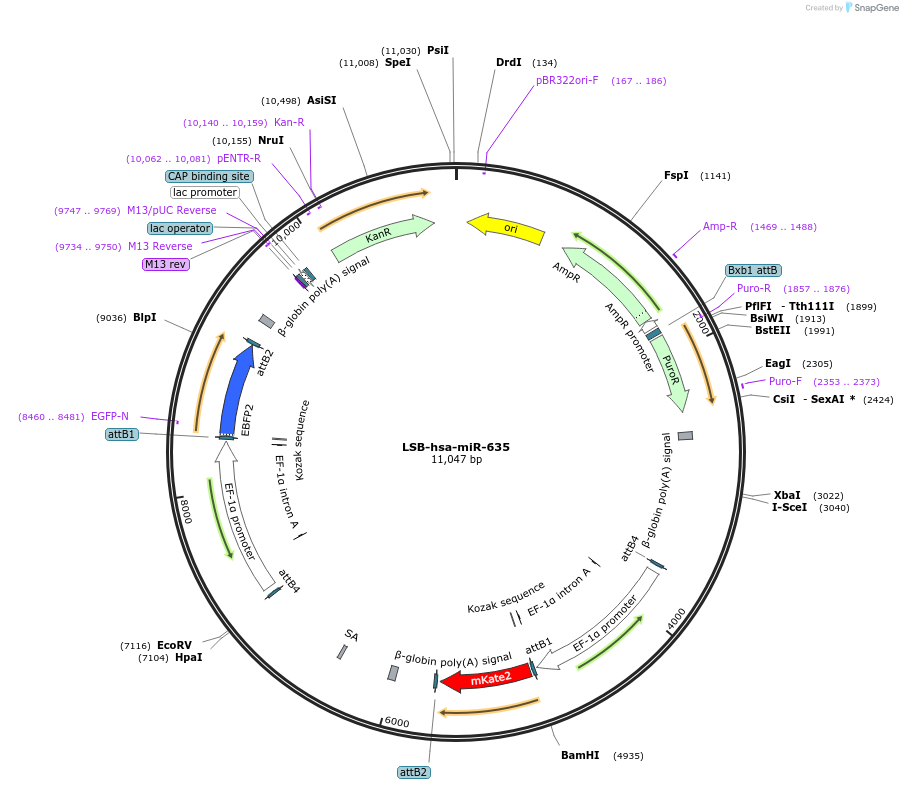
LSB-hsa-miR-635
Plasmid#103693PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-635 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
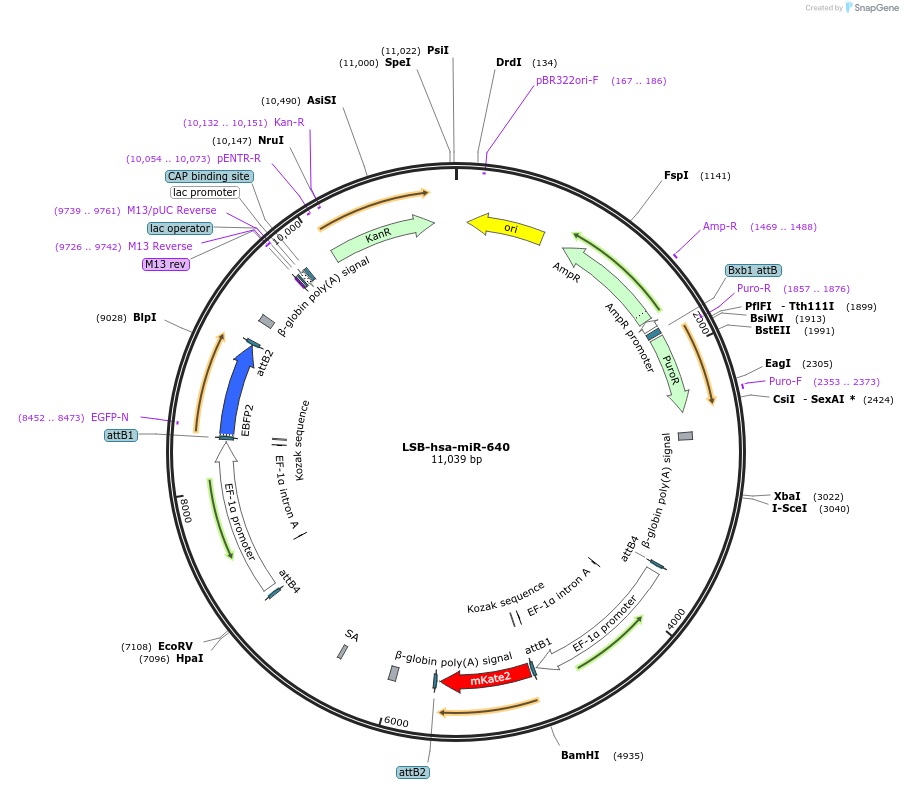
LSB-hsa-miR-640
Plasmid#103695PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-640 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
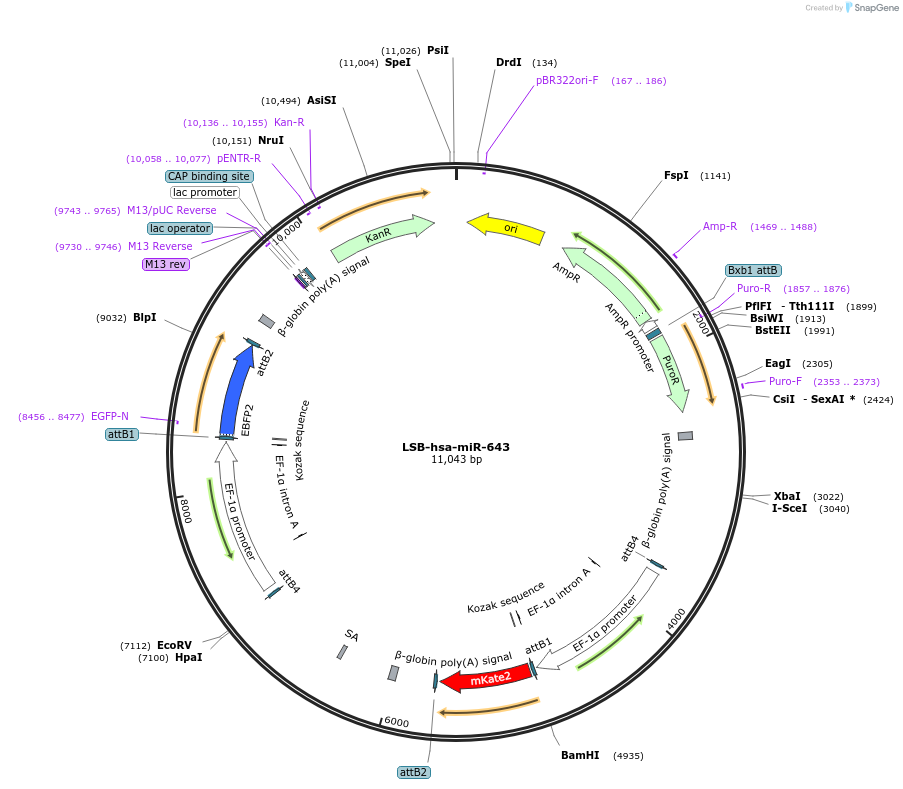
LSB-hsa-miR-643
Plasmid#103698PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-643 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
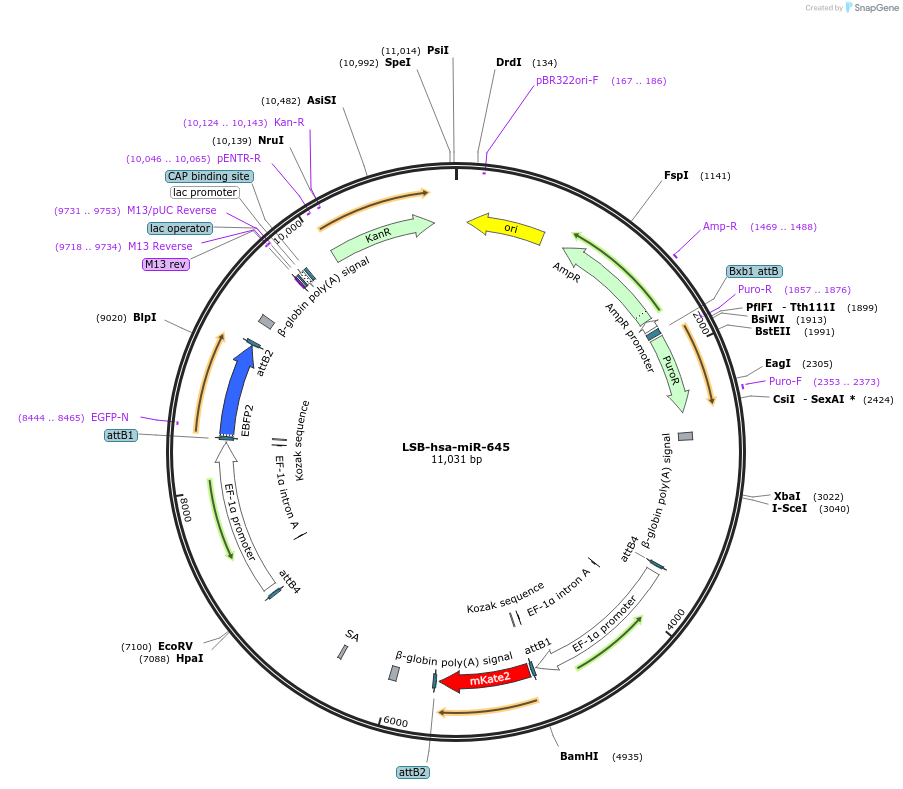
LSB-hsa-miR-645
Plasmid#103699PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-645 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
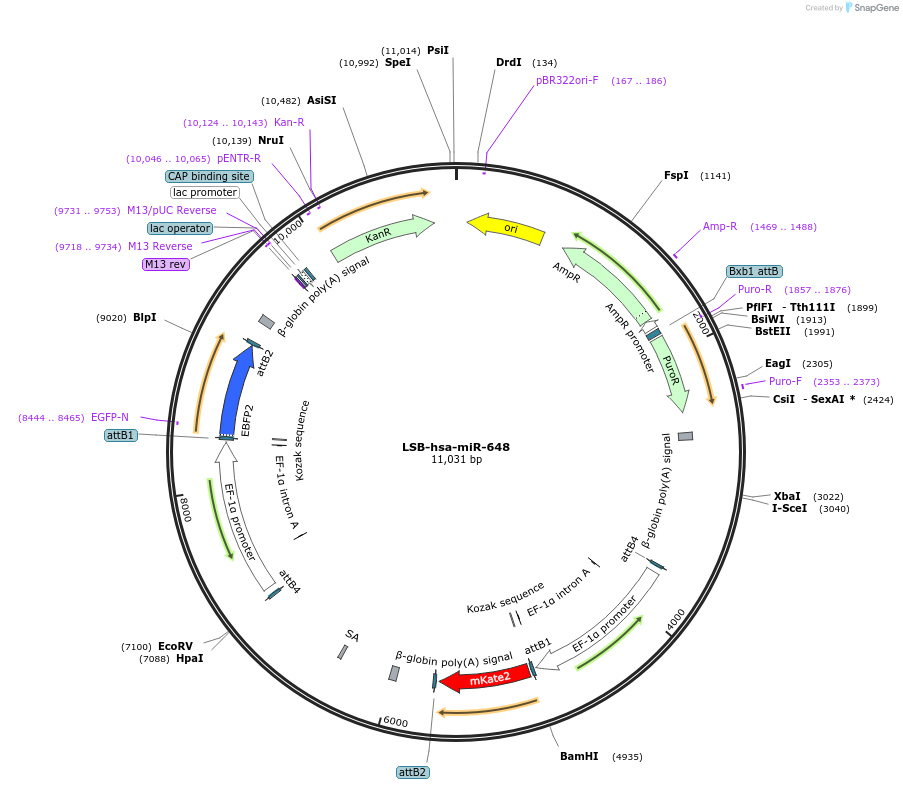
LSB-hsa-miR-648
Plasmid#103700PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-648 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
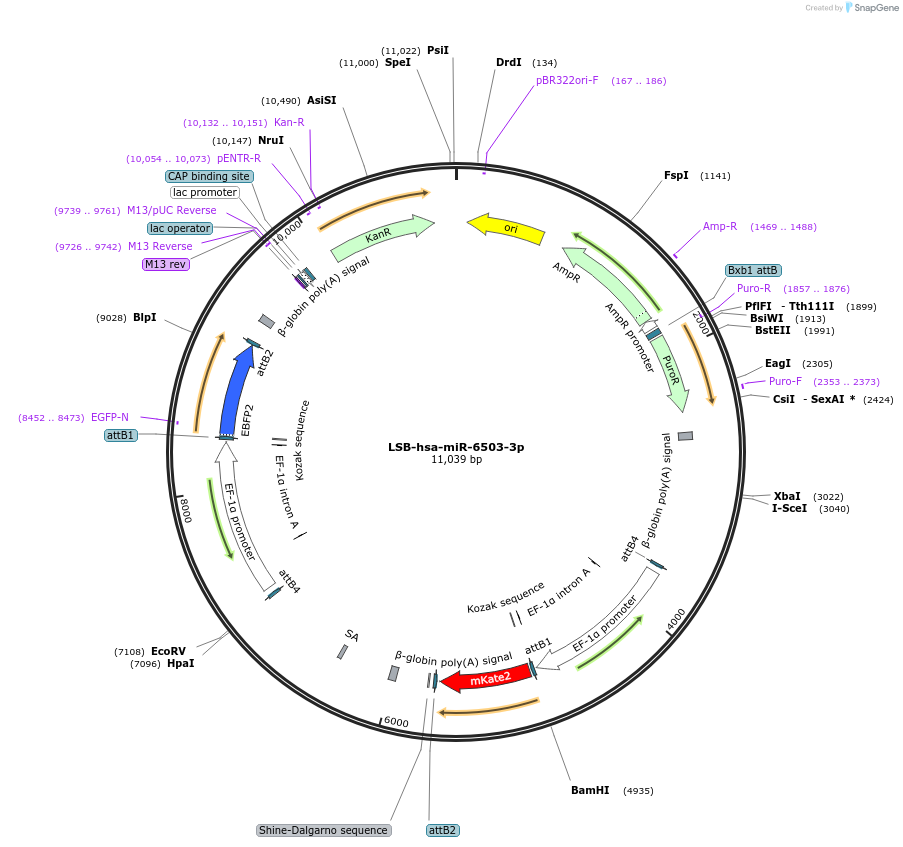
LSB-hsa-miR-6503-3p
Plasmid#103701PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-6503-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-6503-3p target (MIR6503 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
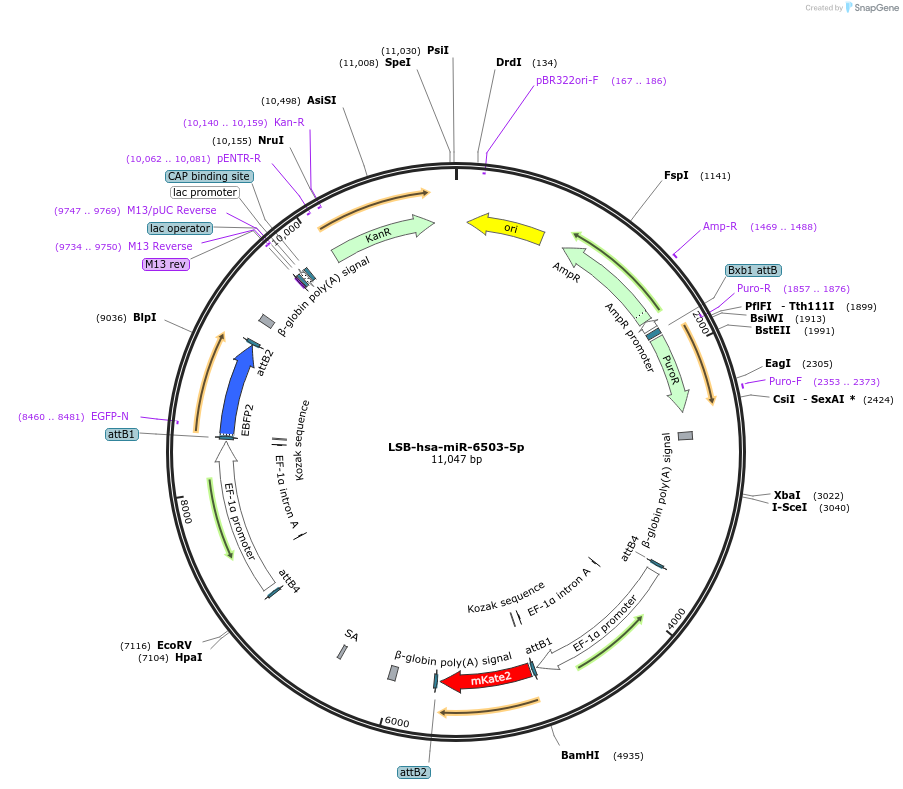
LSB-hsa-miR-6503-5p
Plasmid#103702PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-6503-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-6503-5p target (MIR6503 Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
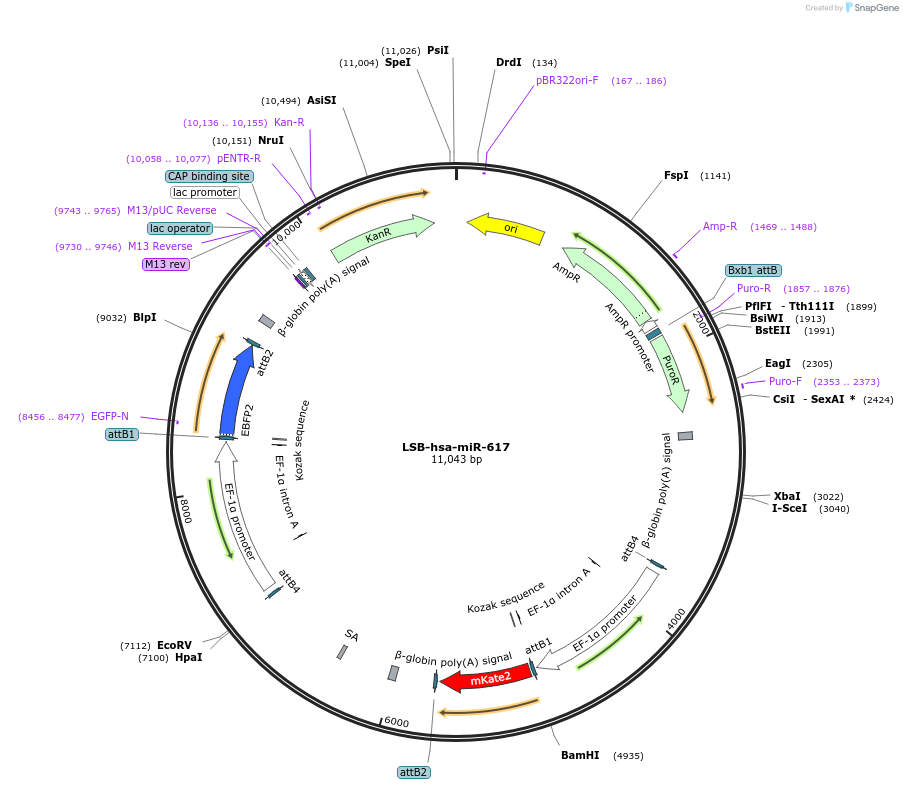
LSB-hsa-miR-617
Plasmid#103678PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-617 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
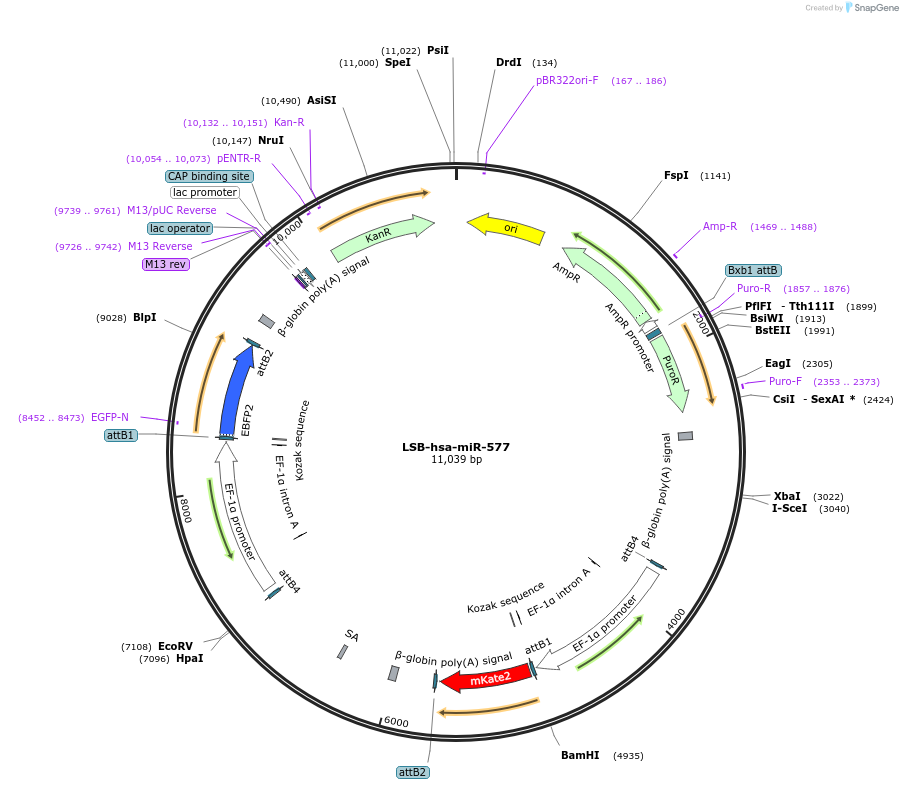
LSB-hsa-miR-577
Plasmid#103659PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-577 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
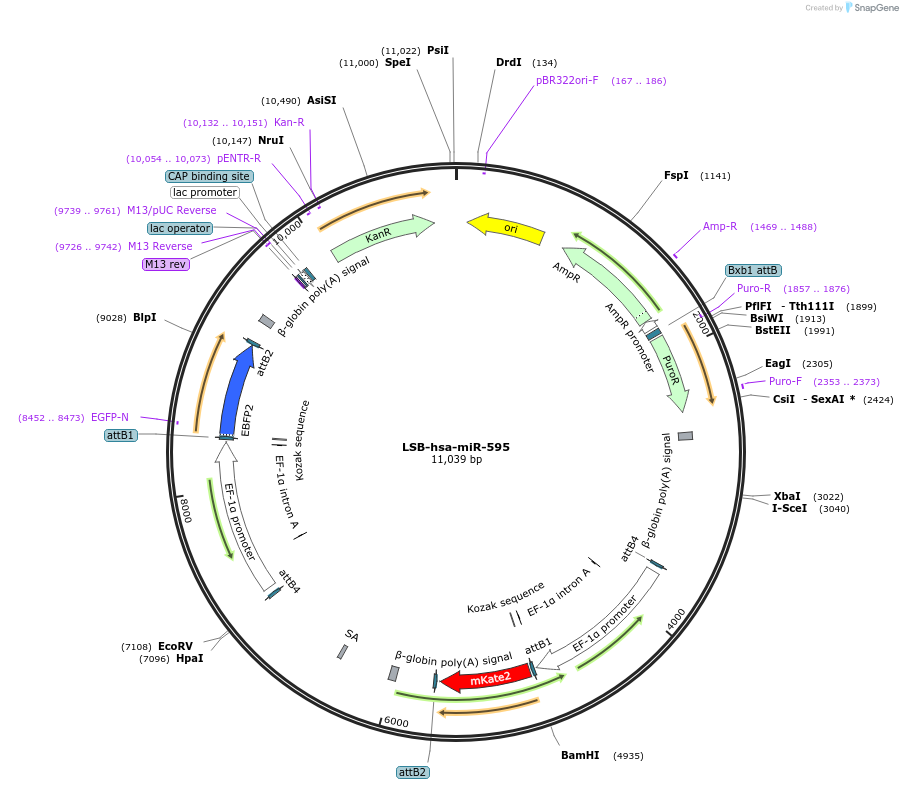
LSB-hsa-miR-595
Plasmid#103670PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-595 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
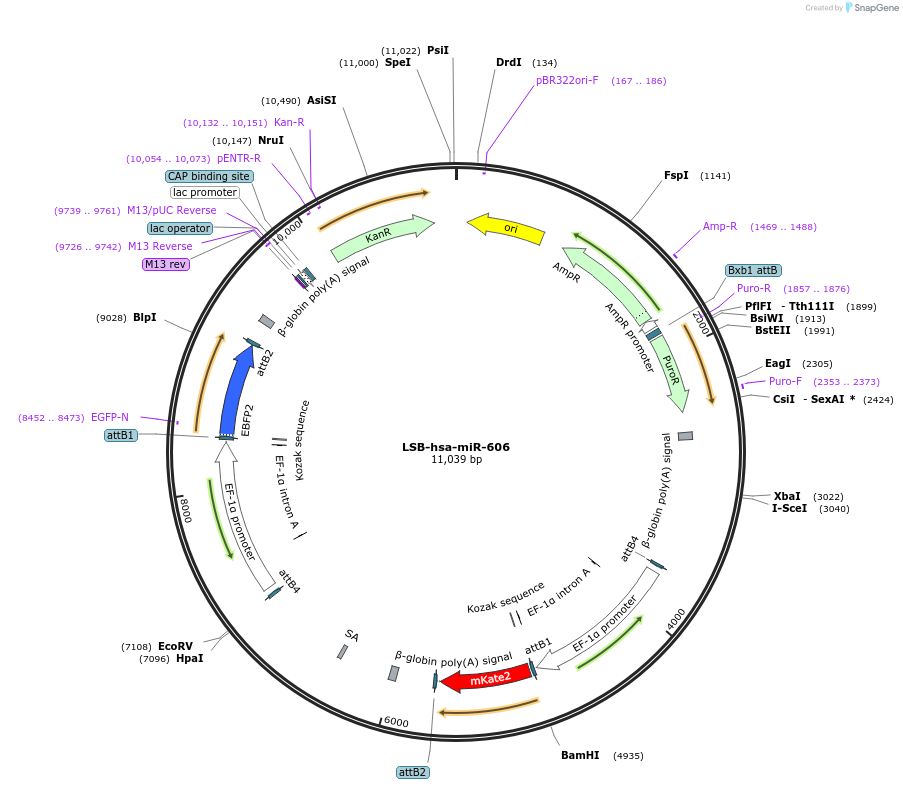
LSB-hsa-miR-606
Plasmid#103671PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-606 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
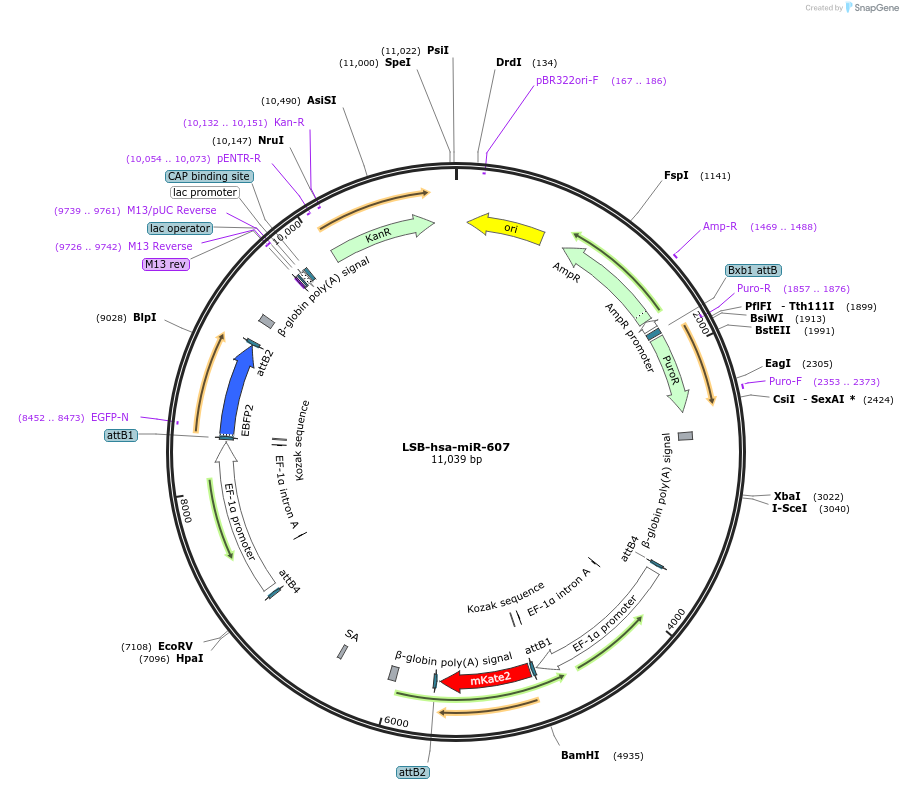
LSB-hsa-miR-607
Plasmid#103672PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-607 target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only