We narrowed to 11,953 results for: SOM
-
Plasmid#158518PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V/L17E-6xHA. Km plant selection marker.DepositorInsertZAR1-6xHA
Tags6xHAExpressionPlantMutationL17E, D481VAvailable SinceNov. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
-
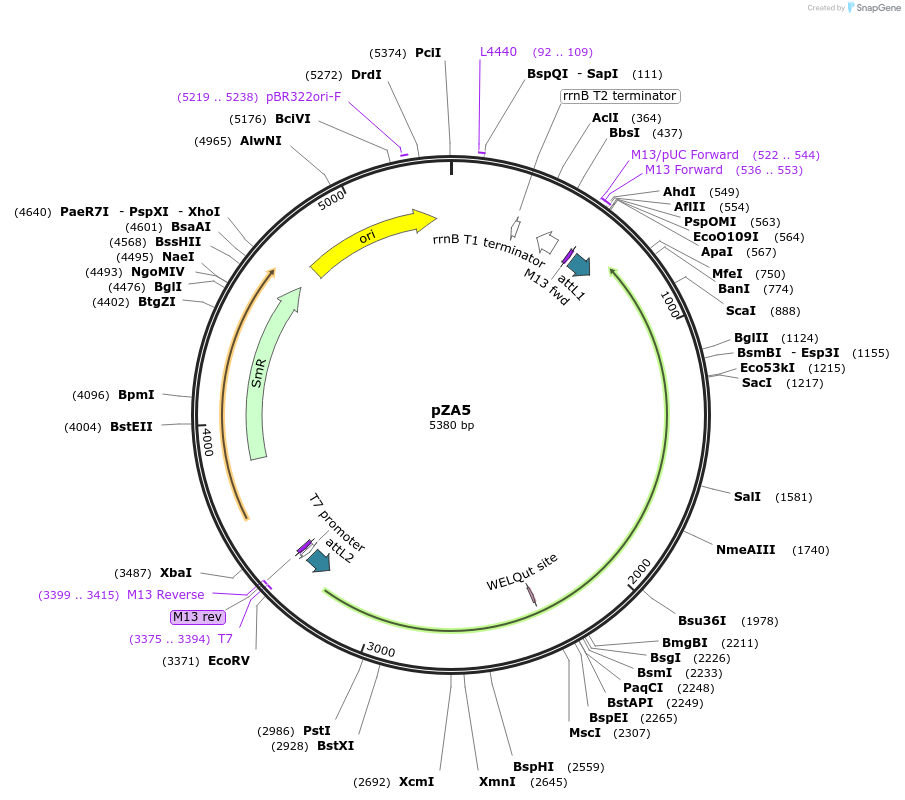
pZA5
Plasmid#158483PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 with STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationSTOP codonAvailable SinceAug. 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
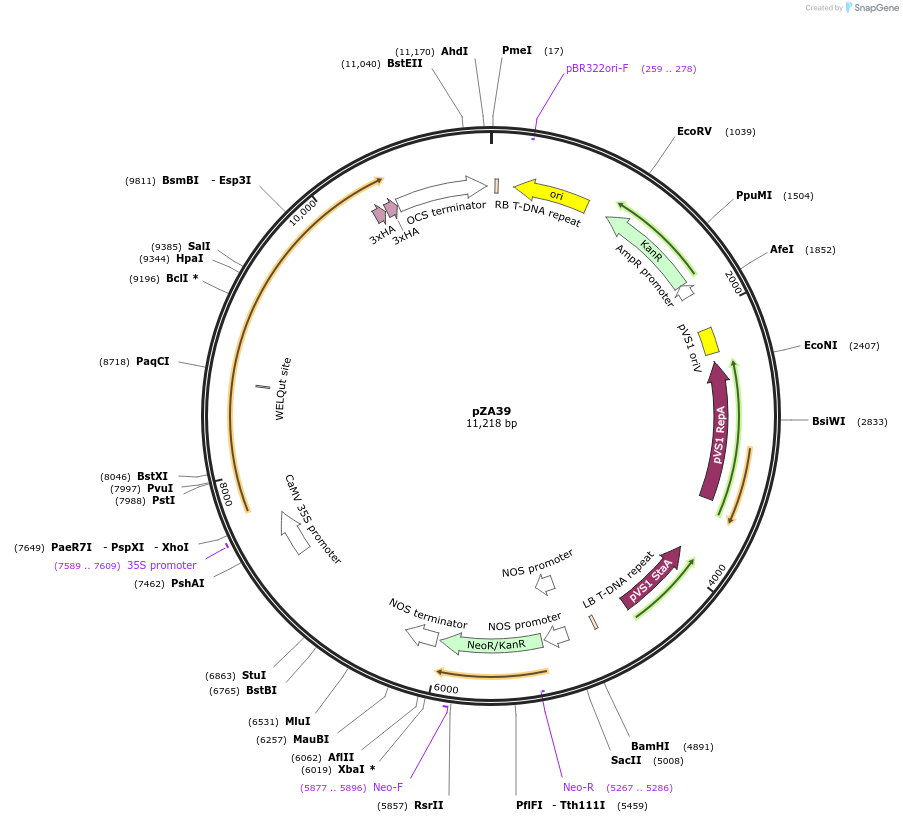
pZA39
Plasmid#158517PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 L17E-6xHA. Km plant selection marker.DepositorInsertZAR1-6xHA
Tags6xHAExpressionPlantMutationL17EAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
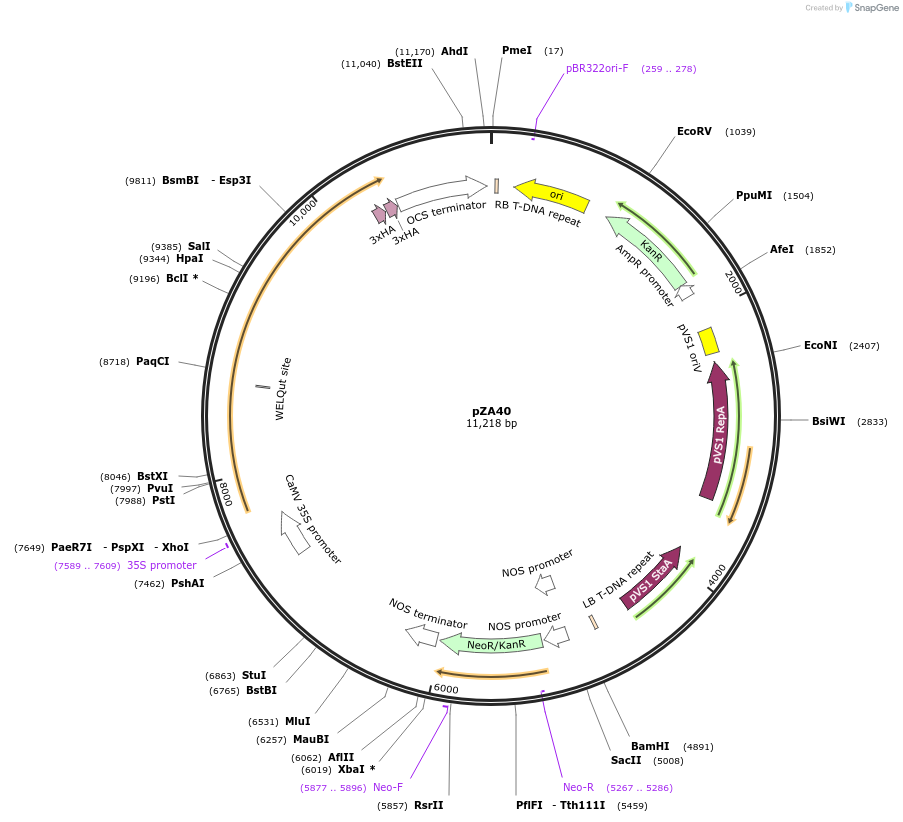
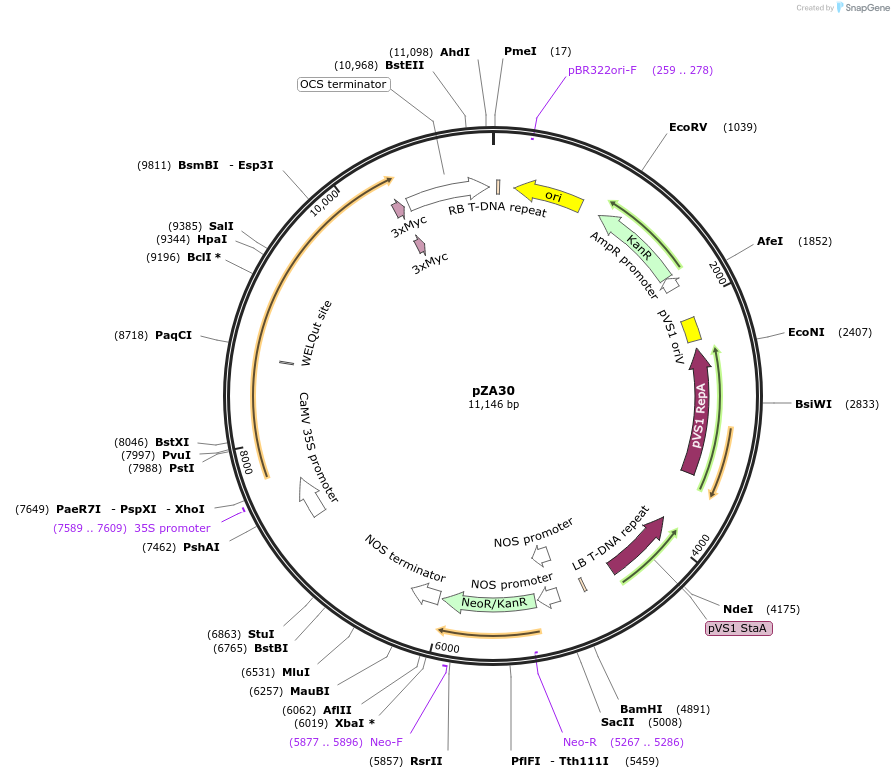
pZA30
Plasmid#158508PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V-4xMyc. Km plant selection marker.DepositorInsertZAR1-4xMyc
Tags4xMycExpressionPlantMutationD481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
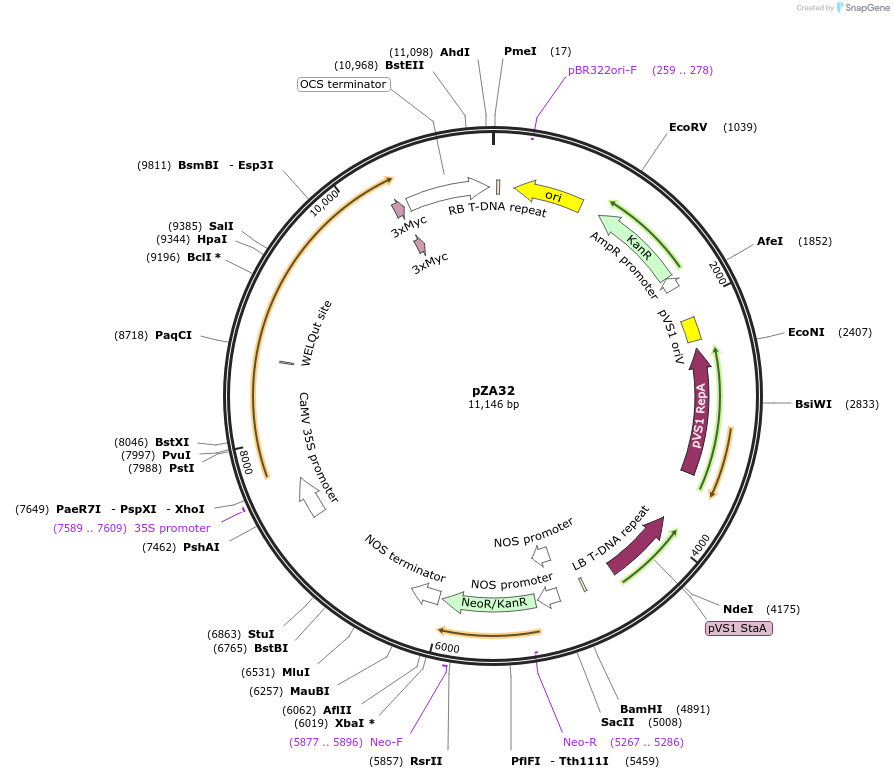
pZA32
Plasmid#158510PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V/L17E-4xMyc. Km plant selection marker.DepositorInsertZAR1-4xMyc
Tags4xMycExpressionPlantMutationL17E, D481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
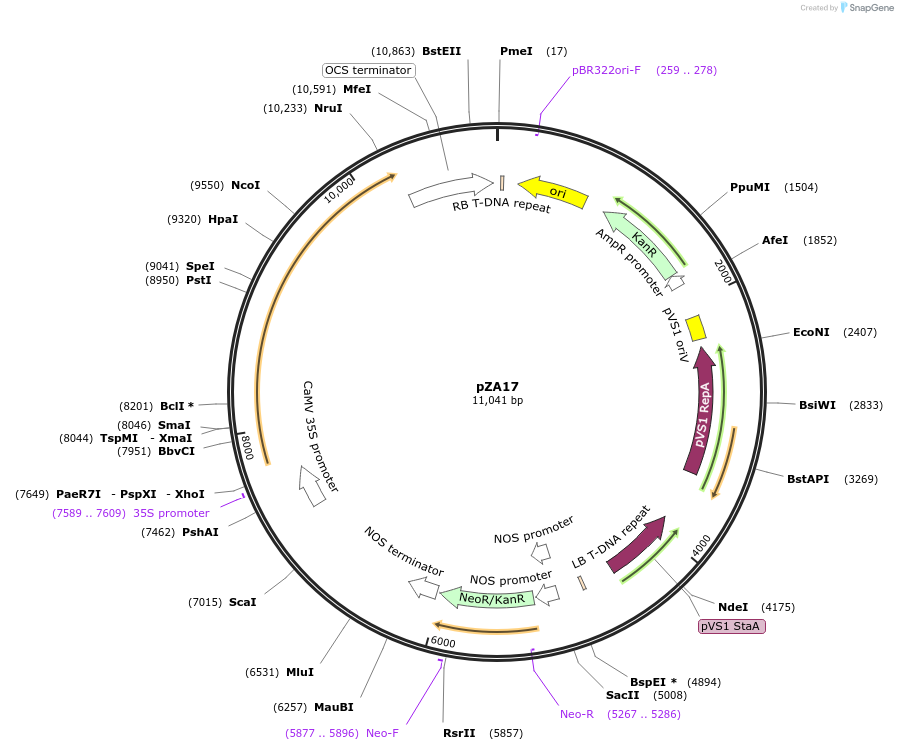
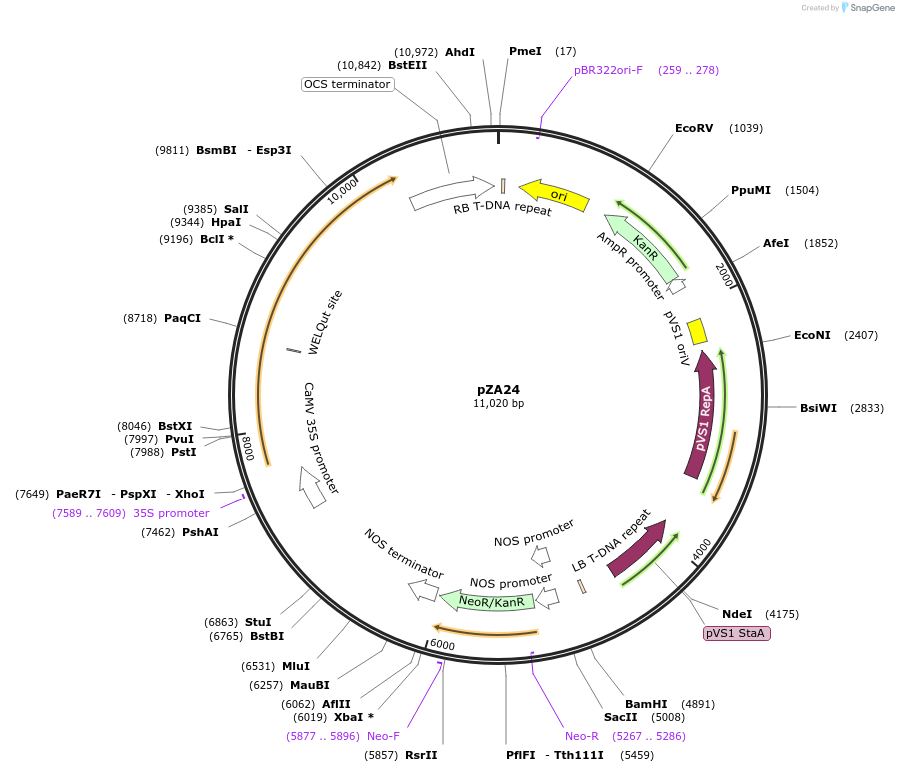
pZA24
Plasmid#158502PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V/L17E. Km plant selection marker.DepositorInsertZAR1
ExpressionPlantMutationL17E, D481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
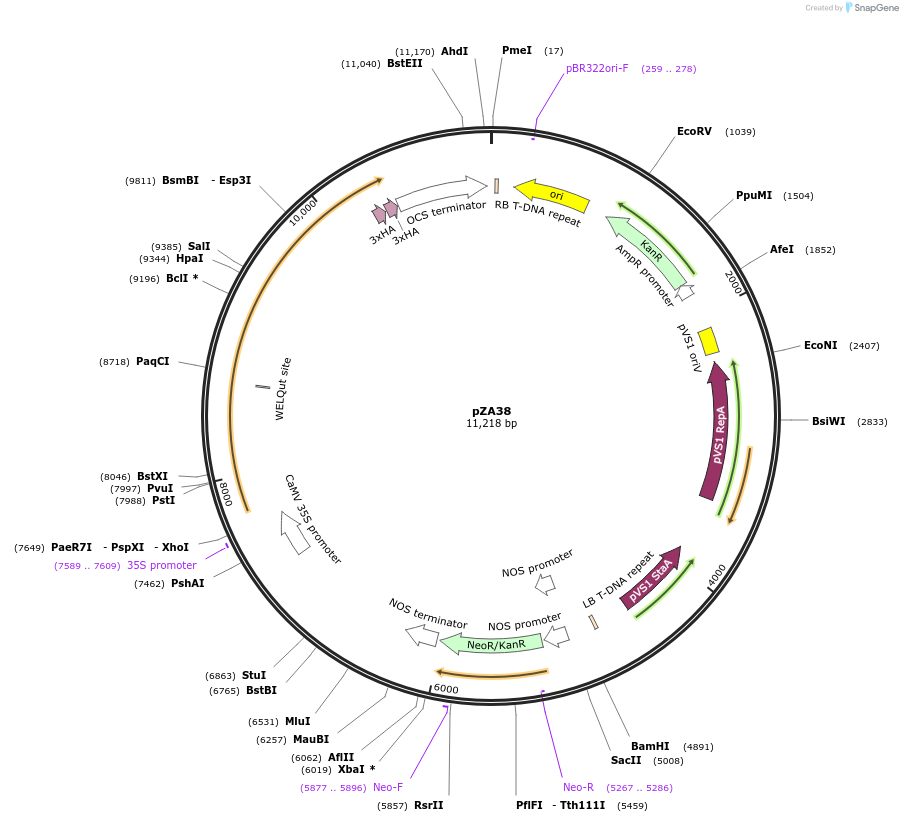
pZA38
Plasmid#158516PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V-6xHA. Km plant selection marker.DepositorInsertZAR1-6xHA
Tags6xHAExpressionPlantMutationD481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
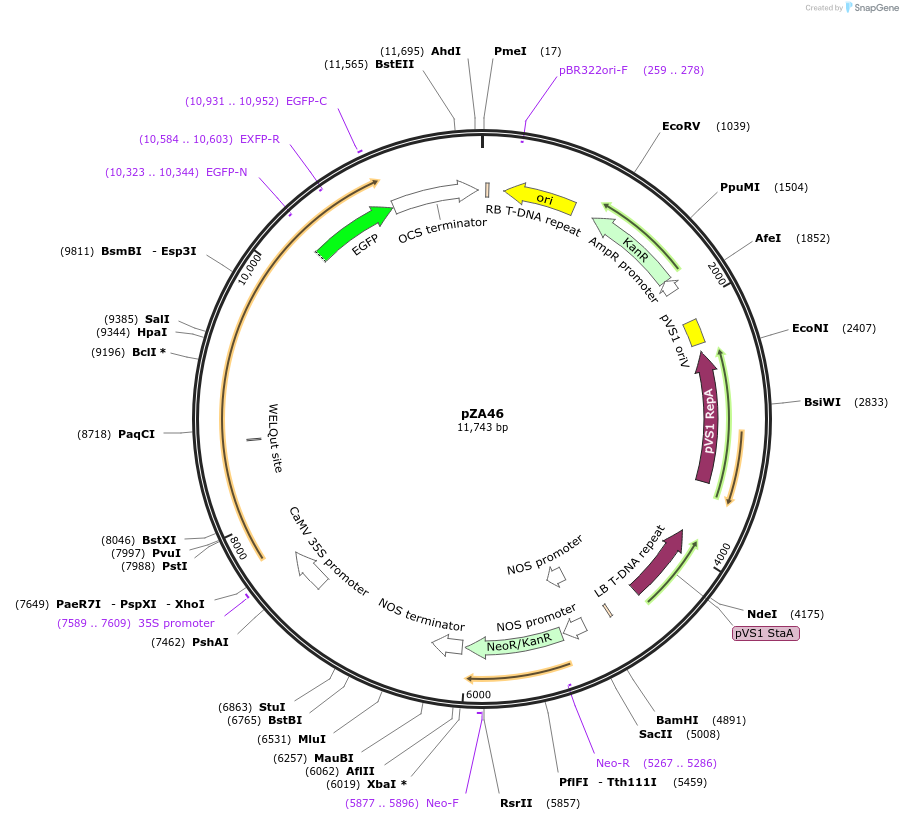
pZA46
Plasmid#158524PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V-eGFP. Km plant selection marker.DepositorInsertZAR1-eGFP
TagseGFPExpressionPlantMutationD481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
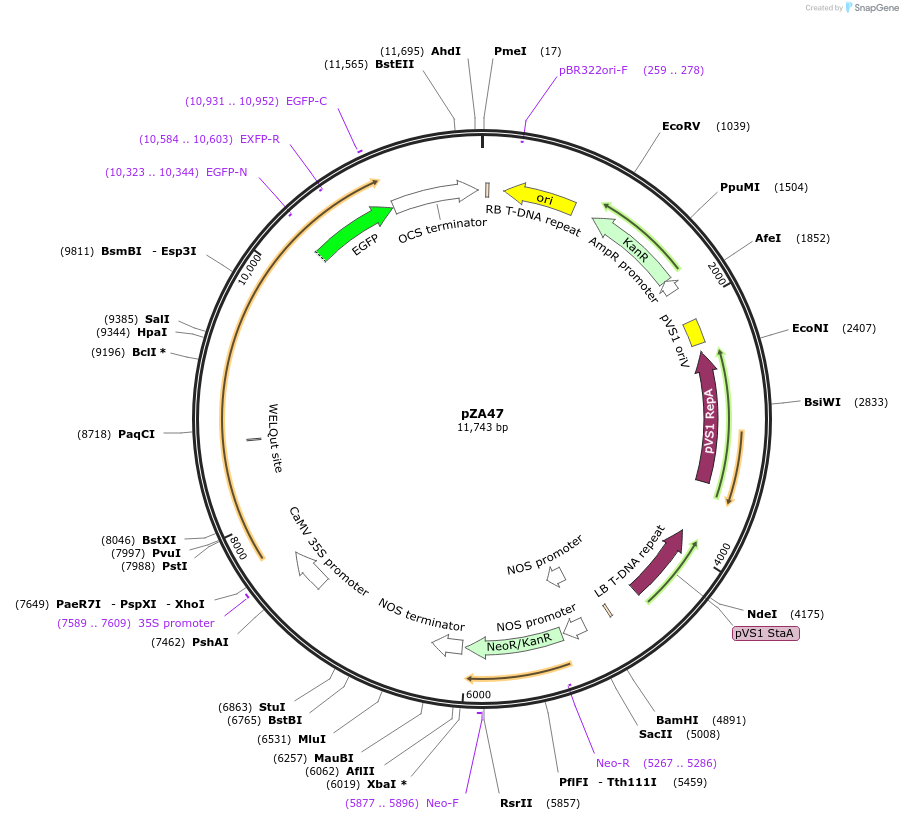
pZA47
Plasmid#158525PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 L17E-eGFP. Km plant selection marker.DepositorInsertZAR1-eGFP
TagseGFPExpressionPlantMutationL17EAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
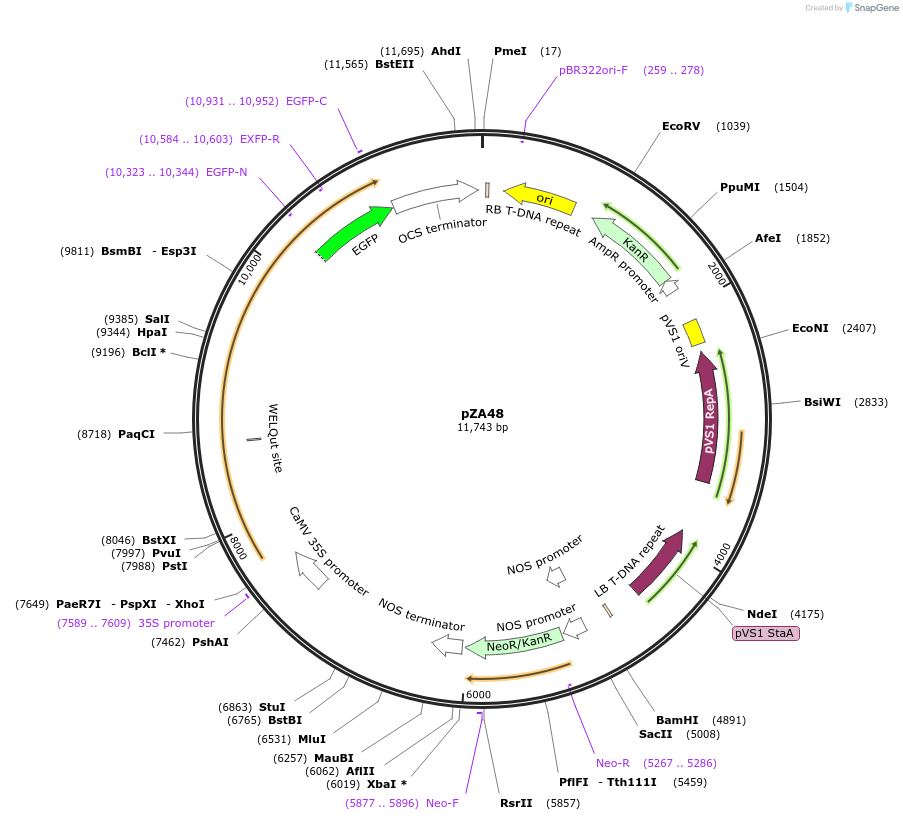
pZA48
Plasmid#158526PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V/L17E-eGFP. Km plant selection marker.DepositorInsertZAR1-eGFP
TagseGFPExpressionPlantMutationL17E, D481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
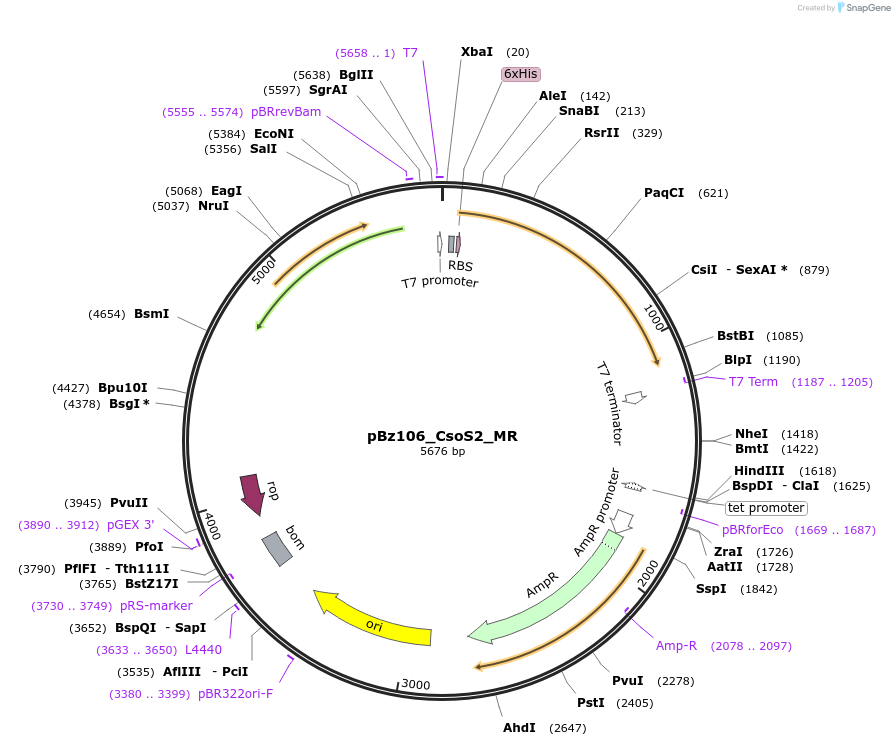
pBz106_CsoS2_MR
Plasmid#140843PurposeHis-tagged CsoS2 Middle Region (MR), pET-14b backboneDepositorInsertCsoS2 Middle Region (MR)
TagsHexahistidine tagExpressionBacterialMutationonly amino acids 257-603PromoterT7Available SinceJune 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
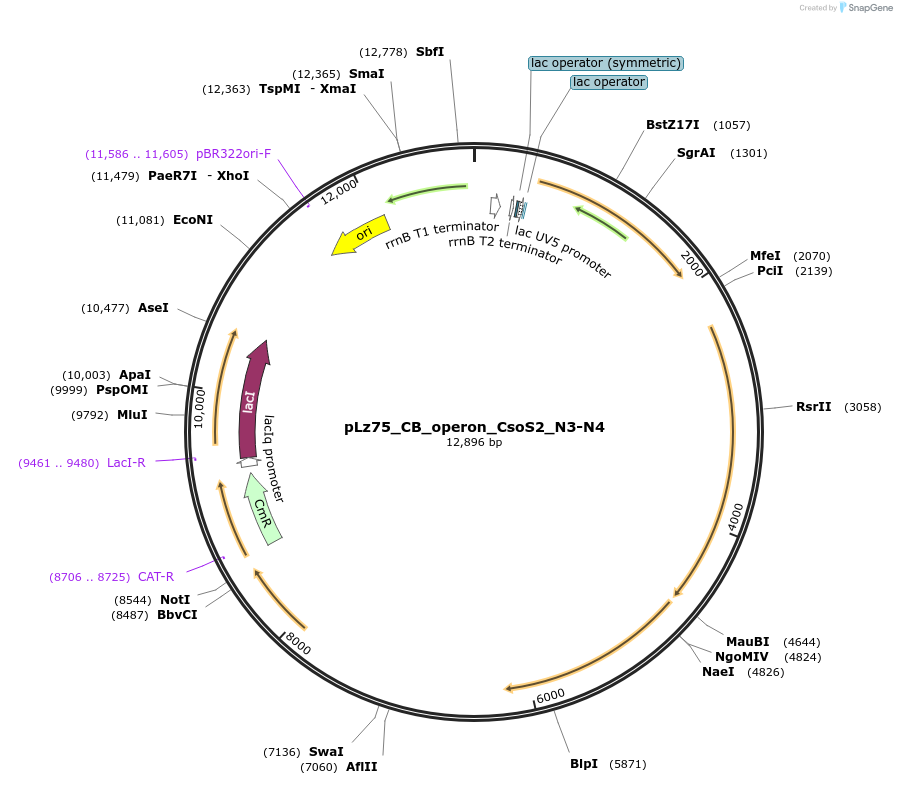
pLz75_CB_operon_CsoS2_N3-N4
Plasmid#140857PurposeSame as pLz37 but with CsoS2 truncated such that NTD repeats N1 and N2 are removedDepositorInsertCbbL, CbbS, CsoS2 (delN1-N2), CsoSCA, CsoS4A, CsoS4B, CsoS1C, CsoS1A, CsoS1B, CsoS1D
ExpressionBacterialMutationdeleted amino acids 1-109 of CsoS2Available SinceJune 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
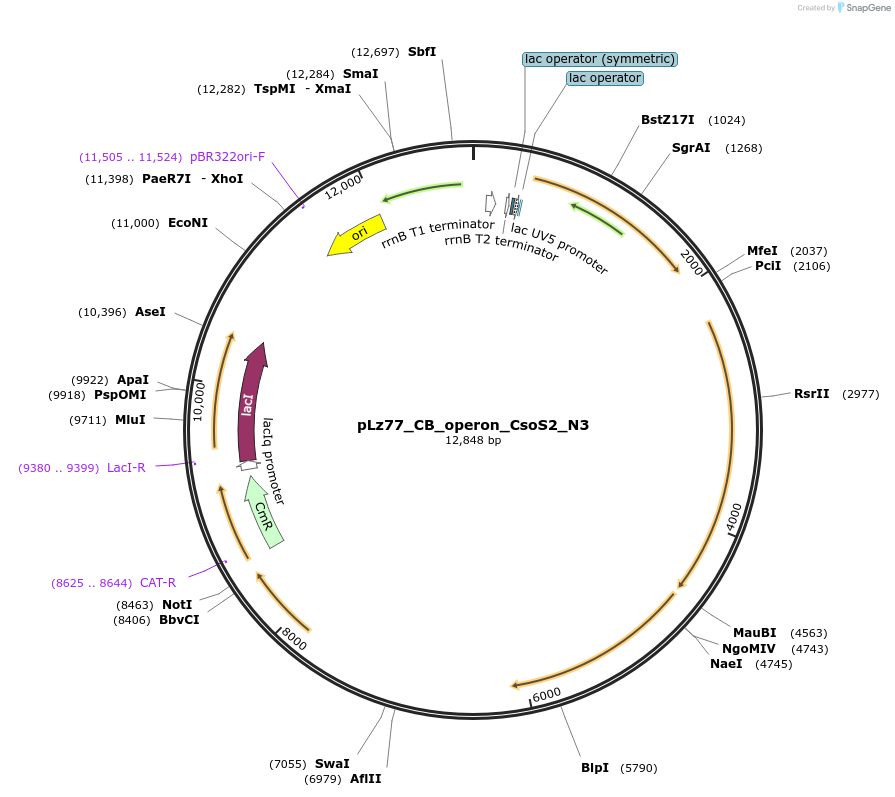
pLz77_CB_operon_CsoS2_N3
Plasmid#140859PurposeSame as pLz37 but with CsoS2 discontinuously truncated such that repeats N1, N2, and N4 are removedDepositorInsertCbbL, CbbS, CsoS2 (delN1-N2, N4), CsoSCA, CsoS4A, CsoS4B, CsoS1C, CsoS1A, CsoS1B, CsoS1D
ExpressionBacterialMutationdeleted amino acids 1-108 and 220-235 of CsoS2Available SinceJune 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
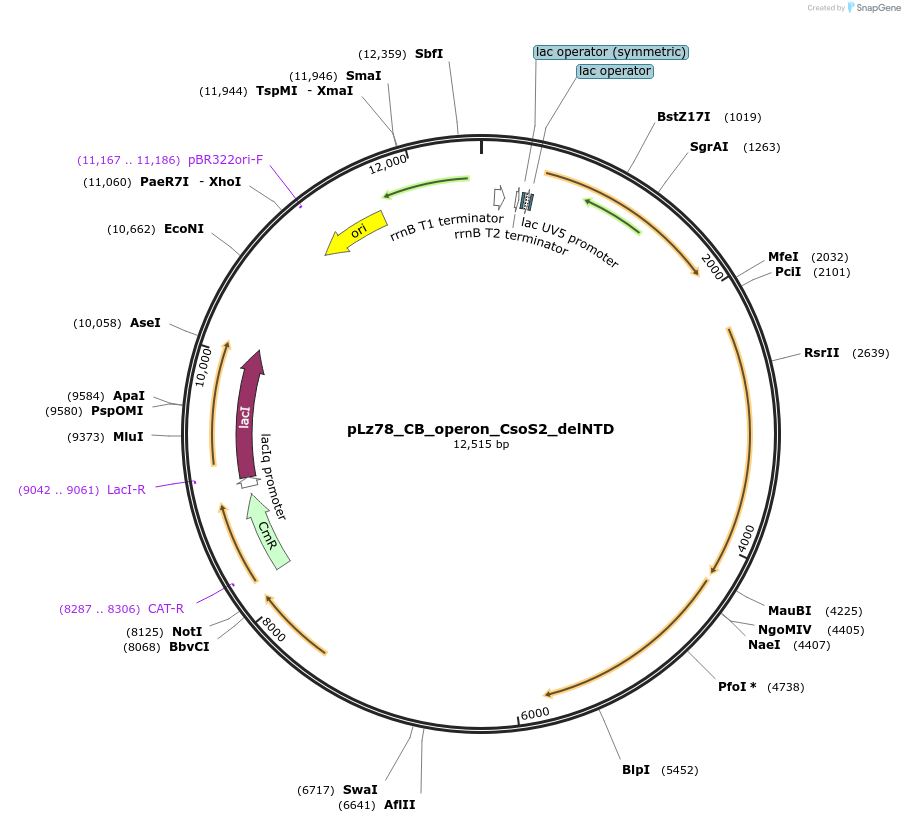
pLz78_CB_operon_CsoS2_delNTD
Plasmid#140860PurposeSame as pLz37 but with CsoS2 truncated to remove the entire NTD (i.e. N1, N2, N3, and N4 are removed)DepositorInsertCbbL, CbbS, CsoS2 (delNTD), CsoSCA, CsoS4A, CsoS4B, CsoS1C, CsoS1A, CsoS1B, CsoS1D
ExpressionBacterialMutationdeleted amino acids 1-235 of CsoS2Available SinceJune 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
pLz76_CB_operon_CsoS2_N4
Plasmid#140858PurposeSame as pLz37 but with CsoS2 truncated such that NTD repeats N1, N2, and N3 are removedDepositorInsertCbbL, CbbS, CsoS2 (delN1-N3), CsoSCA, CsoS4A, CsoS4B, CsoS1C, CsoS1A, CsoS1B, CsoS1D
ExpressionBacterialMutationdeleted amino acids 1-189 of CsoS2Available SinceJune 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
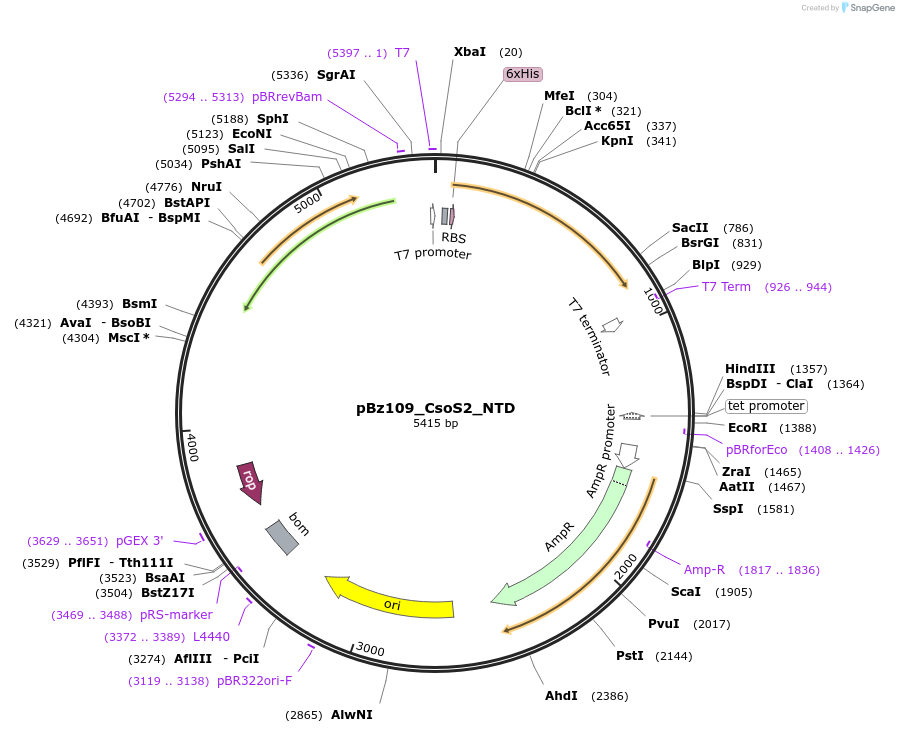
pBz109_CsoS2_NTD
Plasmid#140842PurposeHis-tagged CsoS2 N-terminal domain (NTD), pET-14b backboneDepositorInsertCsoS2 N-terminal domain (NTD)
TagsHexahistidine tagExpressionBacterialMutationdeleted amino acids 262-869PromoterT7Available SinceJune 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
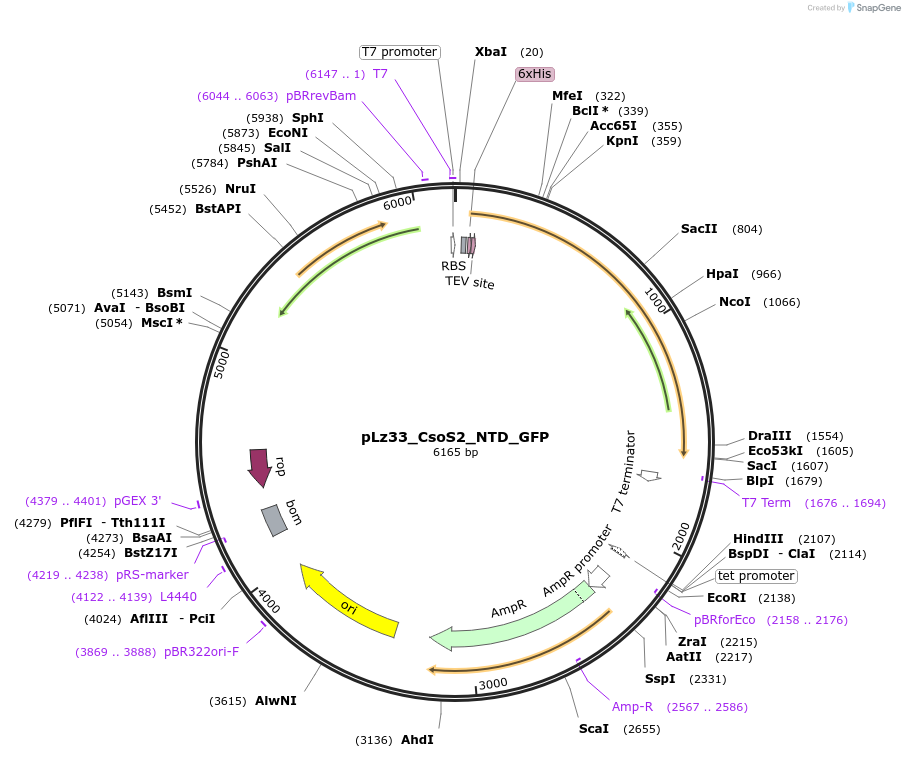
pLz33_CsoS2_NTD_GFP
Plasmid#140861PurposeHis-tagged CsoS2 N-terminal domain (NTD) fused to Superfolder GFPDepositorInsertCsoS2 NTD fused to GFP
TagsHexahistidine tag and Superfolder GFPExpressionBacterialPromoterT7Available SinceMay 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
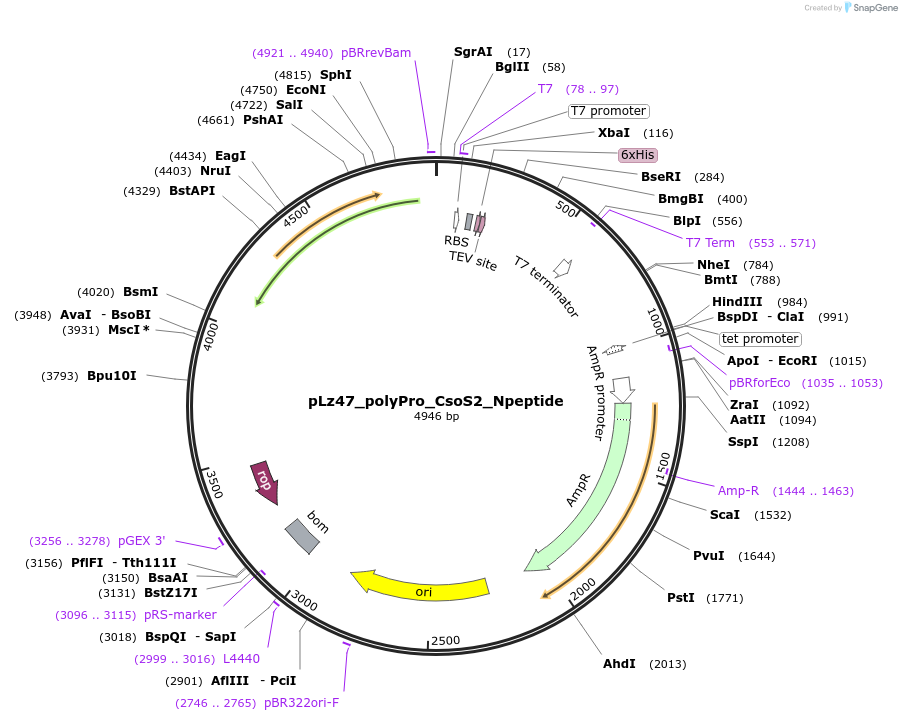
pLz47_polyPro_CsoS2_Npeptide
Plasmid#140845PurposeConsensus N*-peptide after polyproline II helix, N-terminal His- tag used as BLI bait, pET-14b backboneDepositorInsertpolyproline helix with CsoS2 N-peptide
TagsHexahistidine tag and Strep-tag IIExpressionBacterialPromoterT7Available SinceMay 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
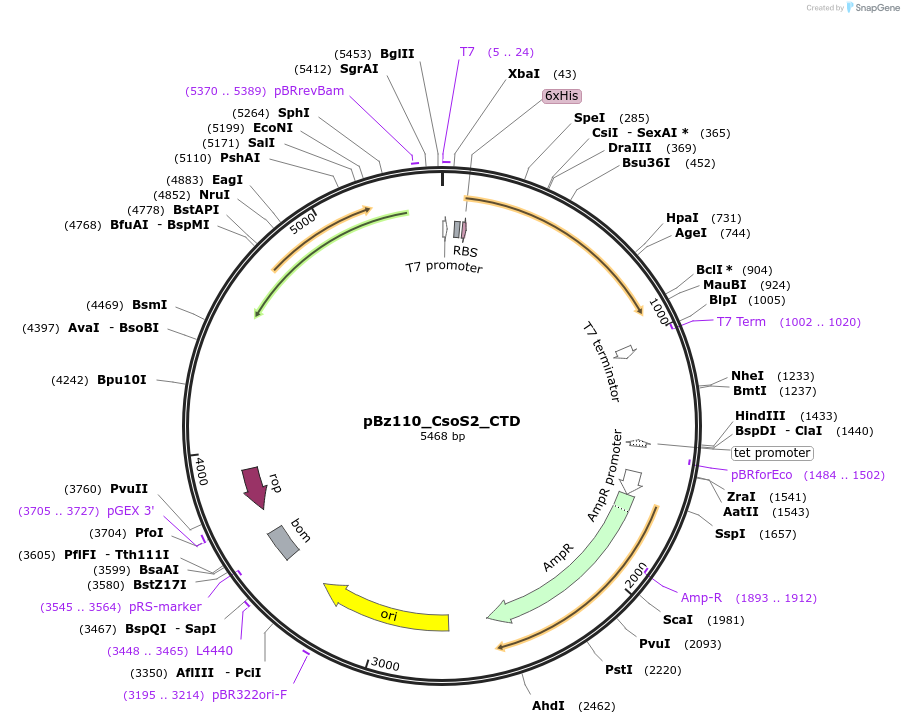
pBz110_CsoS2_CTD
Plasmid#140844PurposeHis-tagged CsoS2 C-terminal domain (CTD), pET-14b backboneDepositorInsertCsoS2 C-terminal domain (CTD)
TagsHexahistidine tagExpressionBacterialMutationdeleted amino acids 1-594PromoterT7Available SinceMay 8, 2020AvailabilityAcademic Institutions and Nonprofits only