We narrowed to 13,064 results for: lic
-
Plasmid#54223PurposeLocalization: Nucleus, Excitation: 487, Emission: 509DepositorAvailable SinceJuly 10, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits
-
mTFP1-H4-23
Plasmid#55491PurposeLocalization: Nucleus/Histones, Excitation: 462, Emission: 492DepositorAvailable SinceOct. 2, 2014AvailabilityAcademic Institutions and Nonprofits only -
mTFP1-Alpha-Actinin-19
Plasmid#55464DepositorAvailable SinceOct. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
pCDF1-ERBB4 (R544W)
Plasmid#29540DepositorAvailable SinceSept. 6, 2011AvailabilityAcademic Institutions and Nonprofits only -
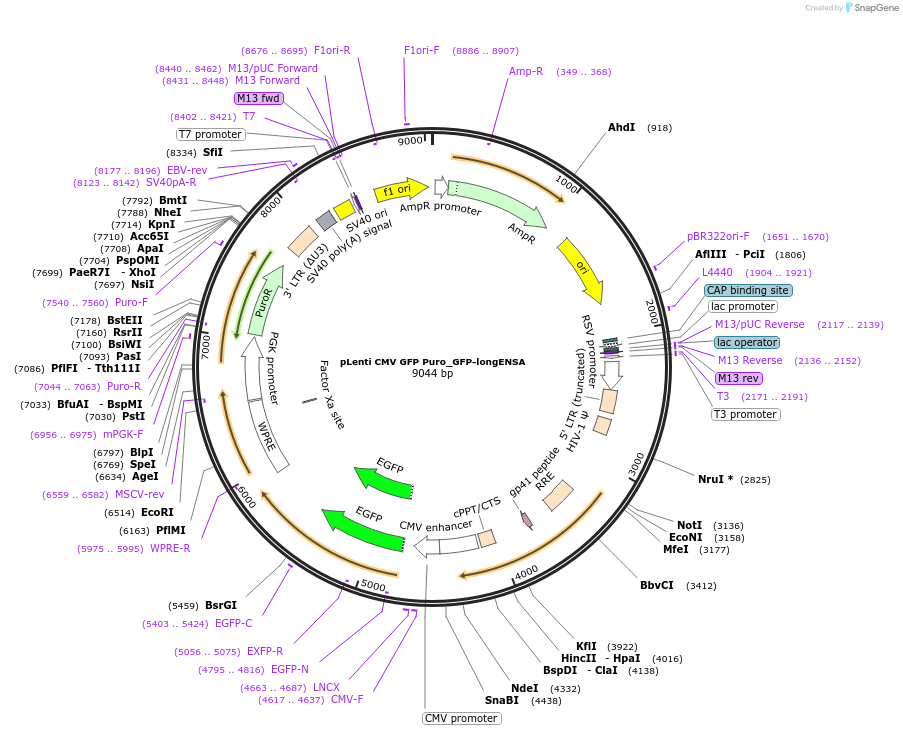
pLenti CMV GFP Puro_GFP-longENSA
Plasmid#248797PurposeOverexpression of long ENSADepositorAvailable SinceDec. 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
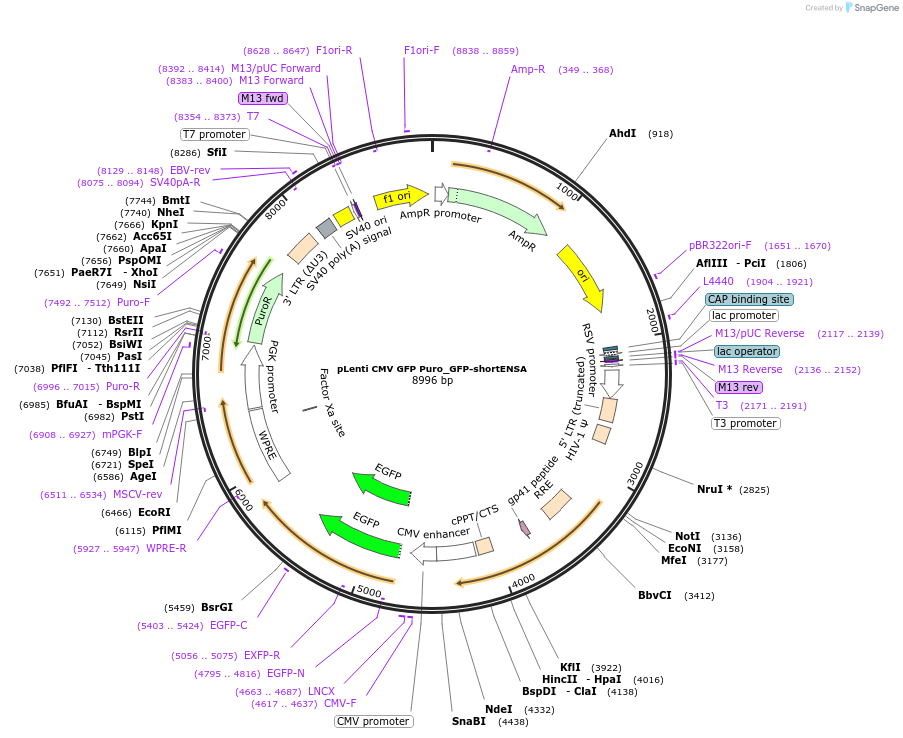
pLenti CMV GFP Puro_GFP-shortENSA
Plasmid#248796PurposeOverexpression of short ENSADepositorAvailable SinceDec. 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
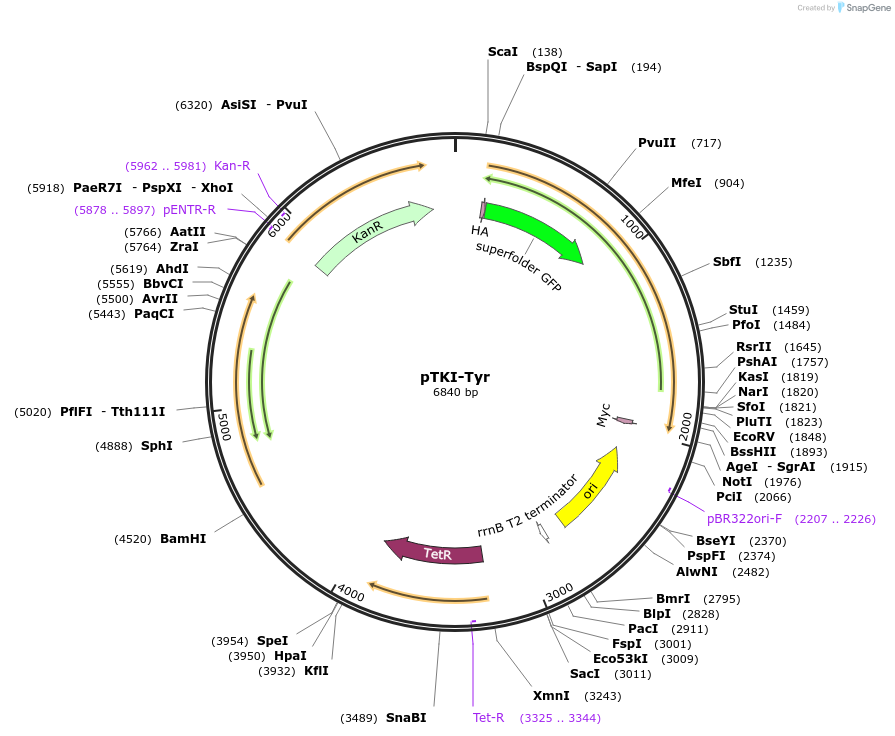
pTKI-Tyr
Plasmid#247647PurposeExpresses GFP-SUMO-Tyr-mCherry N-degron reporter driven by aTc responsive promoter, from KanR integrating mycobacterial plasmid lacking Ulp1DepositorInsertGFP-SUMO-Tyr-mCherry N-degron proteolysis dual-fluorescence reporter
UseMycobacterial integratingTagsHA tag and Myc tagExpressionBacterialPromoterPTetAvailable SinceDec. 9, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
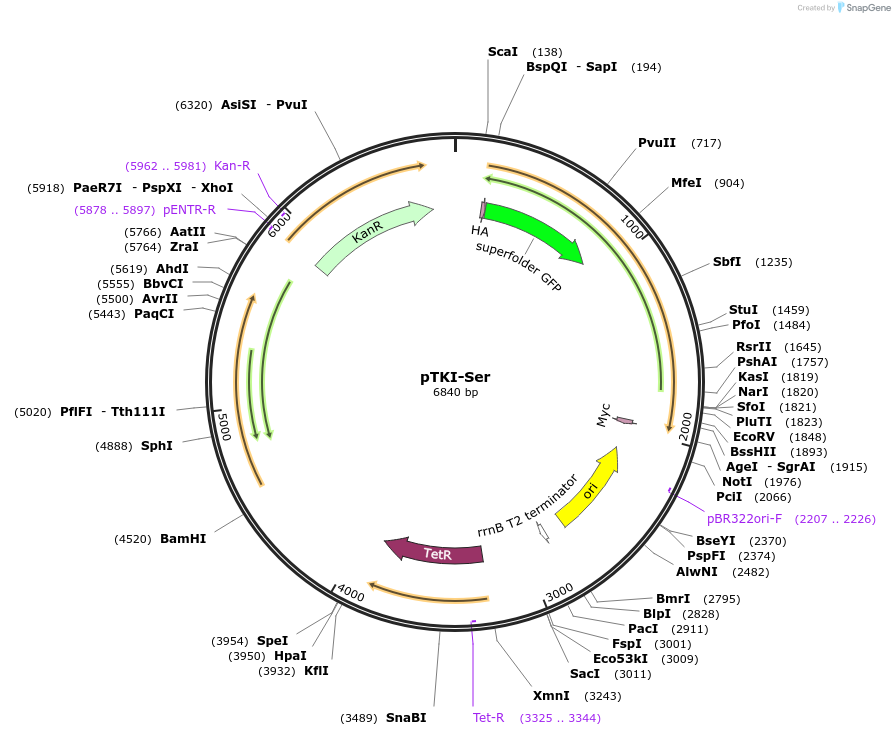
pTKI-Ser
Plasmid#247648PurposeExpresses GFP-SUMO-Ser-mCherry N-degron reporter driven by aTc responsive promoter, from KanR integrating mycobacterial plasmid lacking Ulp1DepositorInsertGFP-SUMO-Ser-mCherry N-degron proteolysis dual-fluorescence reporter
UseMycobacterial integratingTagsHA tag and Myc tagExpressionBacterialPromoterPTetAvailable SinceDec. 9, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
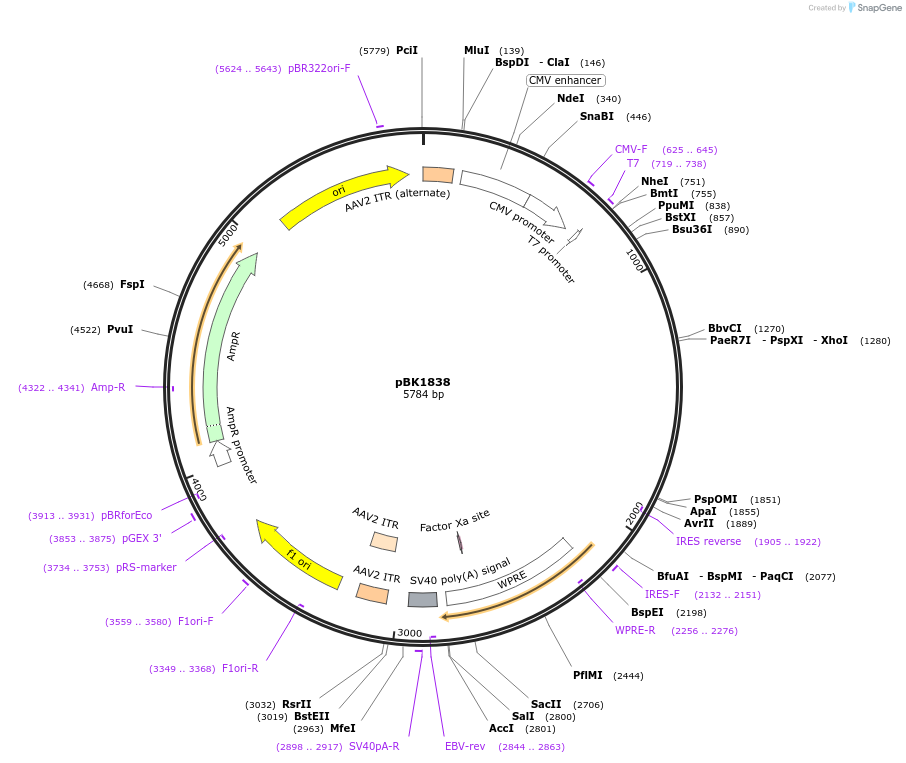
pBK1838
Plasmid#239792PurposepAAV-CMV-SNCAintron1-AlphaSyn[A53T]-WPRE-SV40pA (PD-INDUCER)DepositorInsertCMV/SNCAintron1-AlphaSyn[A53T]-WPRE-SV40pA
UseAAVAvailable SinceNov. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
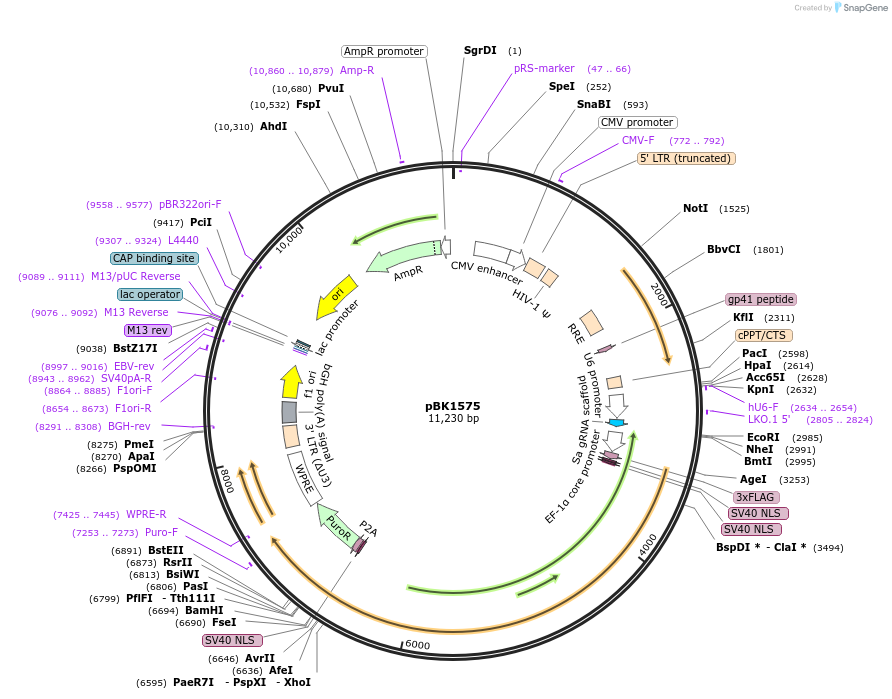
pBK1575
Plasmid#239793PurposepLV-U6-[empty]-EFSNC-dSaCas9-[empty linker]-P2A-Puro-WPREDepositorInsertpLV-U6-[empty]-EFSNC-dSaCas9-[empty linker]-P2A-Puro-WPRE
UseLentiviralAvailable SinceNov. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
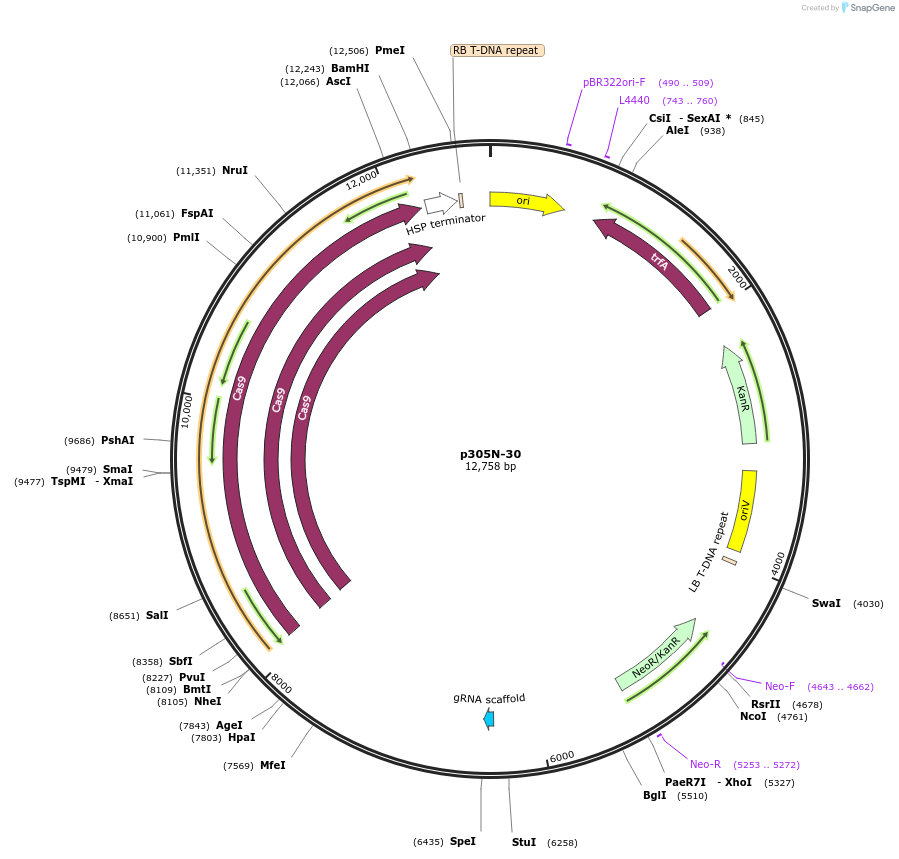
p305N-30
Plasmid#246313PurposeEvaluation of PtU6.2c3 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c3 promoter
UseCRISPRExpressionPlantMutationdeletion: -T (-29 from TSS) within TATAPromoterPtU6.2c3 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
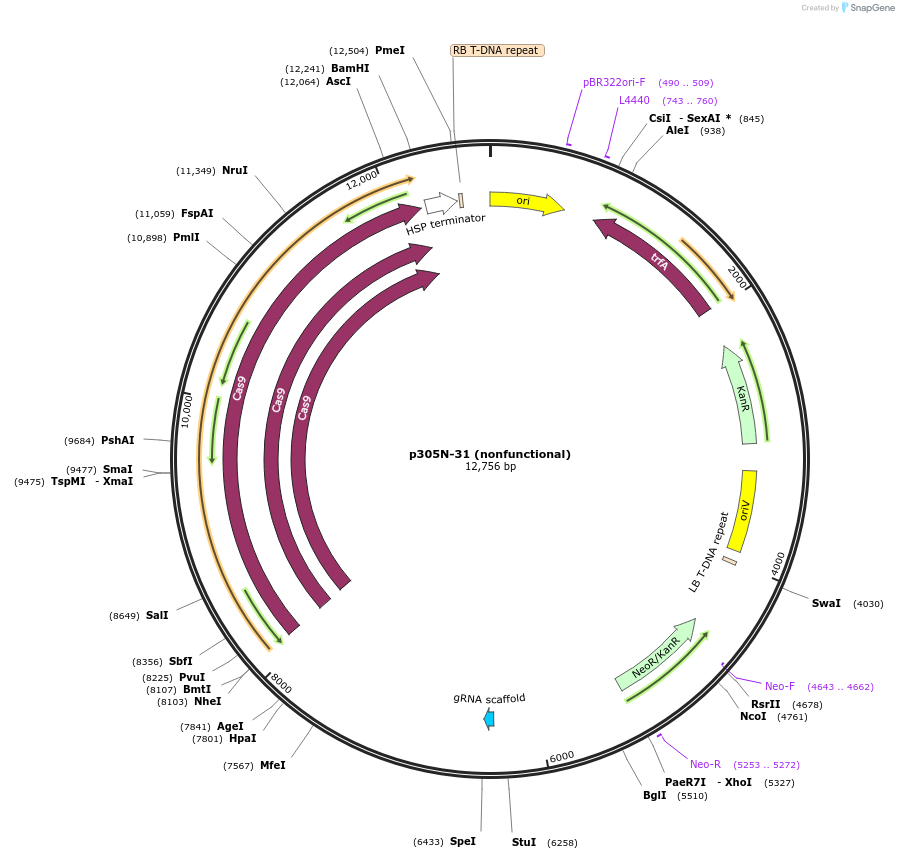
p305N-31 (nonfunctional)
Plasmid#246314PurposeEvaluation of PtU6.2c4 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c4 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationdeletions: -A (-41 from TSS), -T (-30), -T (-29);…PromoterPtU6.2c4 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
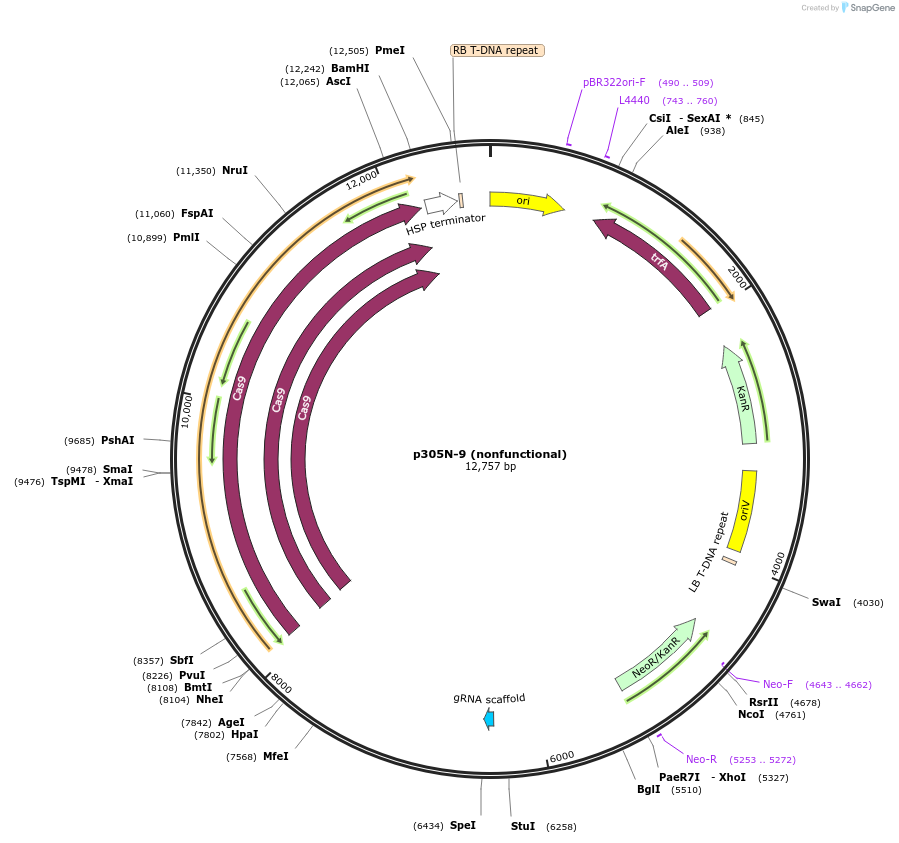
p305N-9 (nonfunctional)
Plasmid#246292PurposeEvaluation of AtU6.29c13 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.29c13 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationdeletions: -A (-58 from TSS), -T (-57) within USEPromoterAtU6.29c13 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
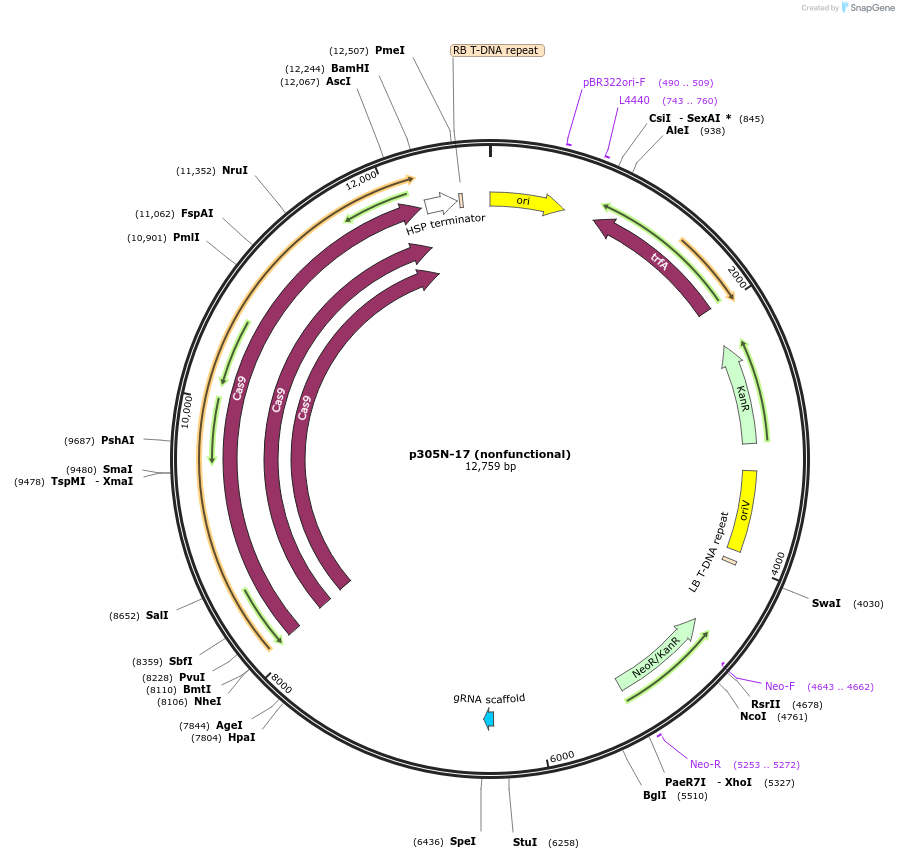
p305N-17 (nonfunctional)
Plasmid#246300PurposeEvaluation of HbU6.2m3 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertHbU6.2m3 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-T (-29 from TSS), G-to-A (-24)PromoterHbU6.2m3 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
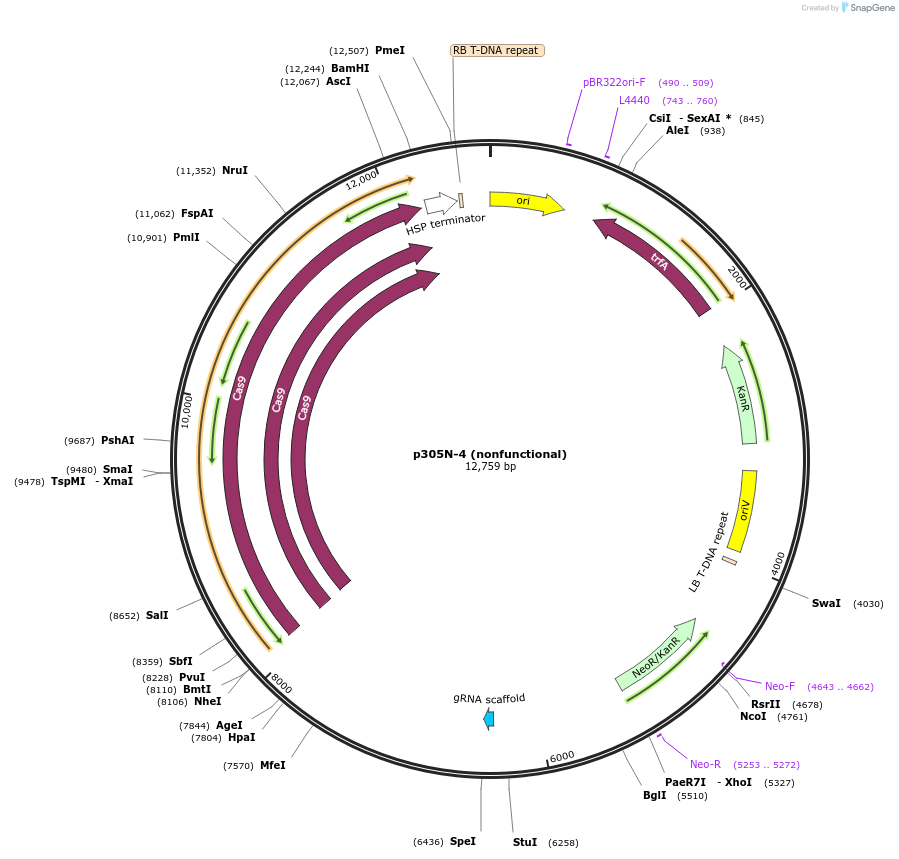
p305N-4 (nonfunctional)
Plasmid#246287PurposeEvaluation of AtU6.1c1 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.1c1 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationG-to-C at -15 from TSSPromoterAtU6.1c1 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
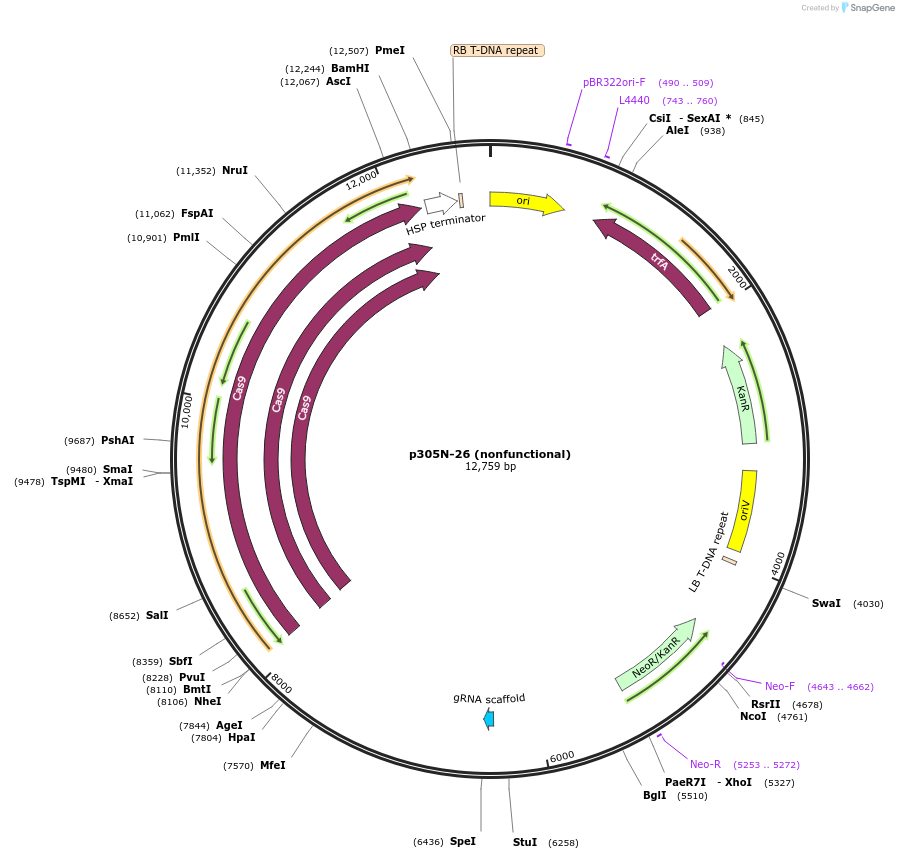
p305N-26 (nonfunctional)
Plasmid#246309PurposeEvaluation of MtU6.6m7 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertMtU6.6m7 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-A (-63 from TSS), T-to-G (-57)PromoterMtU6.6m7 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
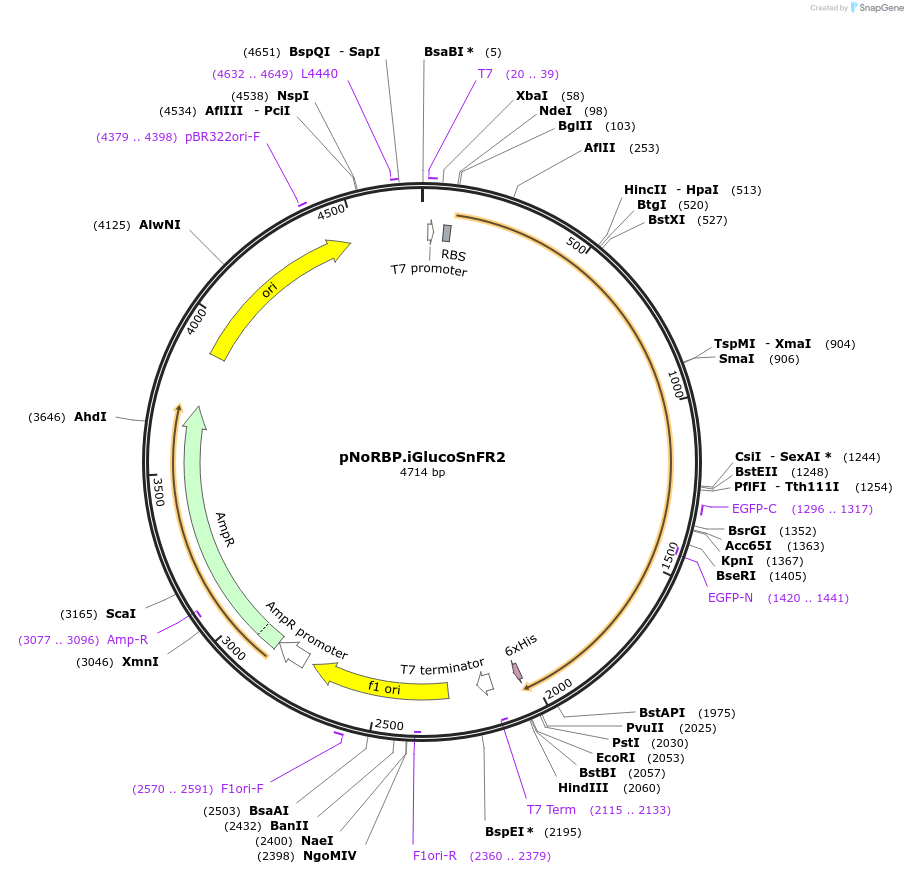
pNoRBP.iGlucoSnFR2
Plasmid#244104PurposeBacterial expression of green glucose sensorDepositorInsertiGlucoSnFR2
ExpressionBacterialAvailable SinceSept. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
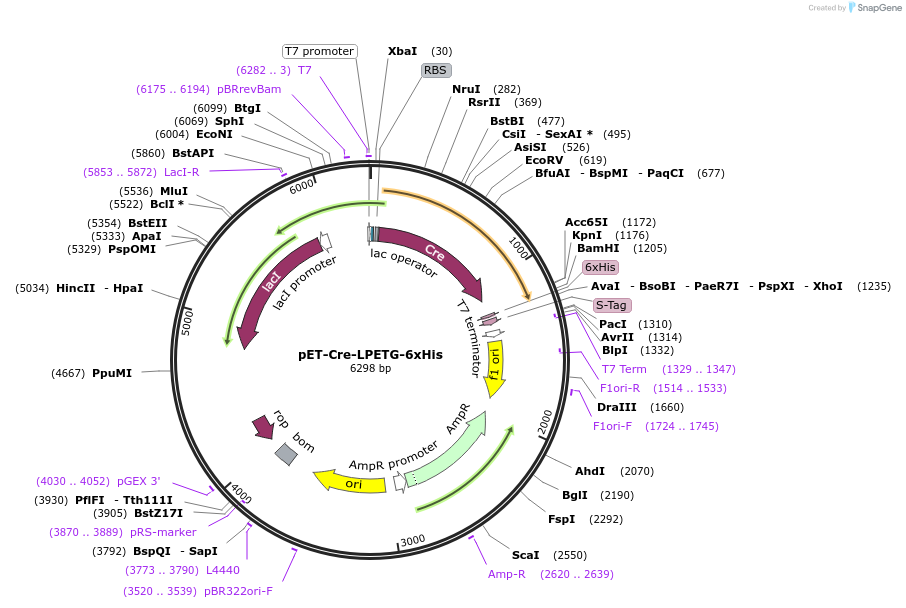
pET-Cre-LPETG-6xHis
Plasmid#237224PurposeExpresses LPETG-tagged Cre recombinase in bacteriaDepositorAvailable SinceJuly 29, 2025AvailabilityAcademic Institutions and Nonprofits only -
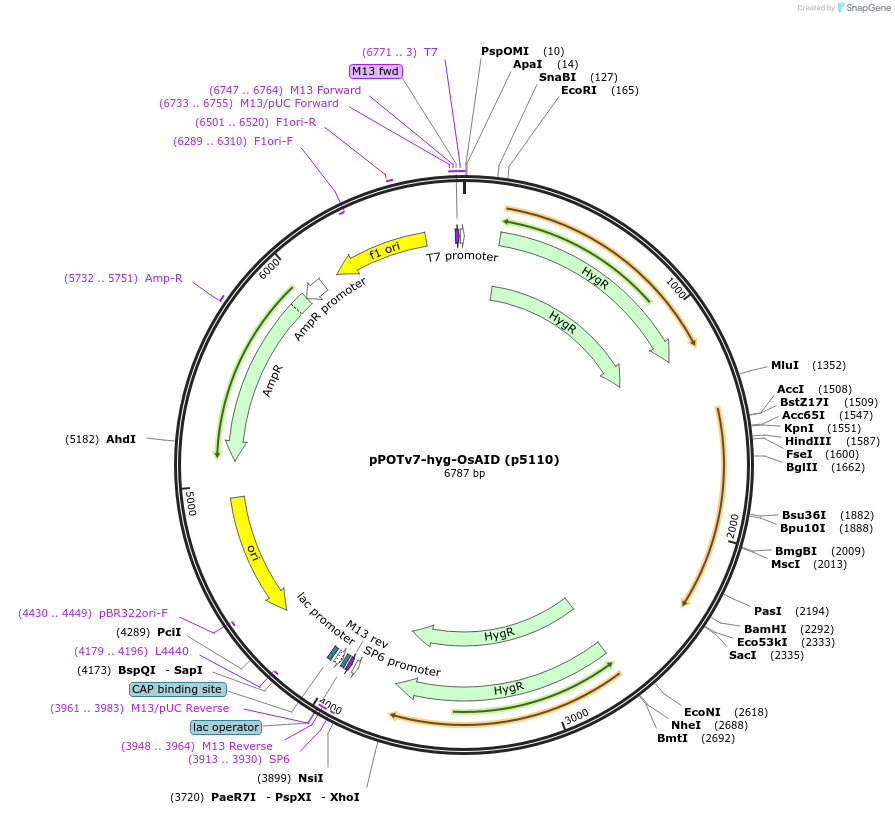
pPOTv7-hyg-OsAID (p5110)
Plasmid#239360PurposepPOTv7 based plasmid template for addition of a Rice Auxin Inducible Degradation (AID) domain to genes in trypanosomes with selection using hygromycinDepositorInsertsRice Auxin Inducible Degradation domain
Auxin Inducible Degradation domain
UseBacterialPromoternoneAvailable SinceJuly 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
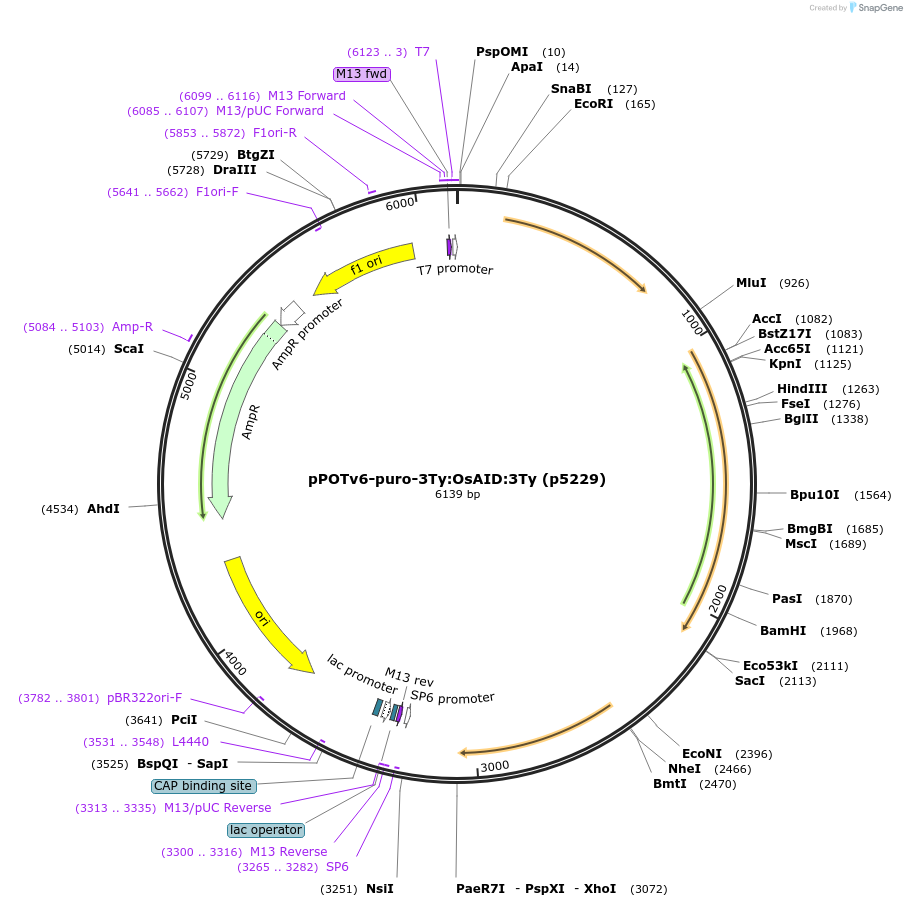
pPOTv6-puro-3Ty:OsAID:3Ty (p5229)
Plasmid#239361PurposepPOTv6 based plasmid template for addition of Ty-epitope tagged Rice Auxin Inducible Degradation (AID) domain to genes in trypanosomes with selection using puromycinDepositorInsertRice Auxin inducible degradation domain
UseBacterialTags3 x Ty epitope tagPromoternoneAvailable SinceJuly 16, 2025AvailabilityAcademic Institutions and Nonprofits only