We narrowed to 2,122 results for: Rp1;
-
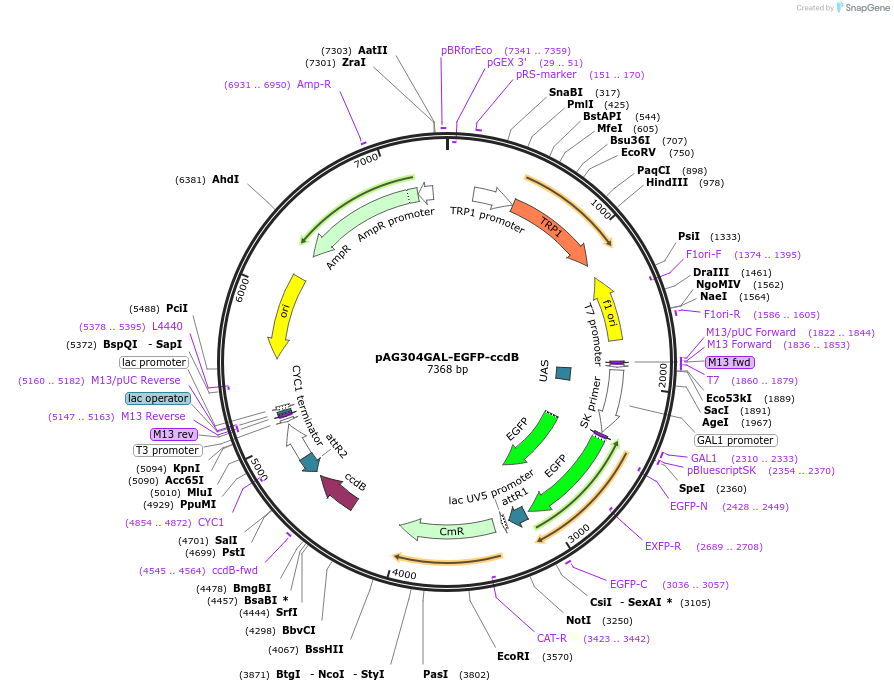
Plasmid#14303DepositorTypeEmpty backboneUseGateway destinationTagsEGFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only
-
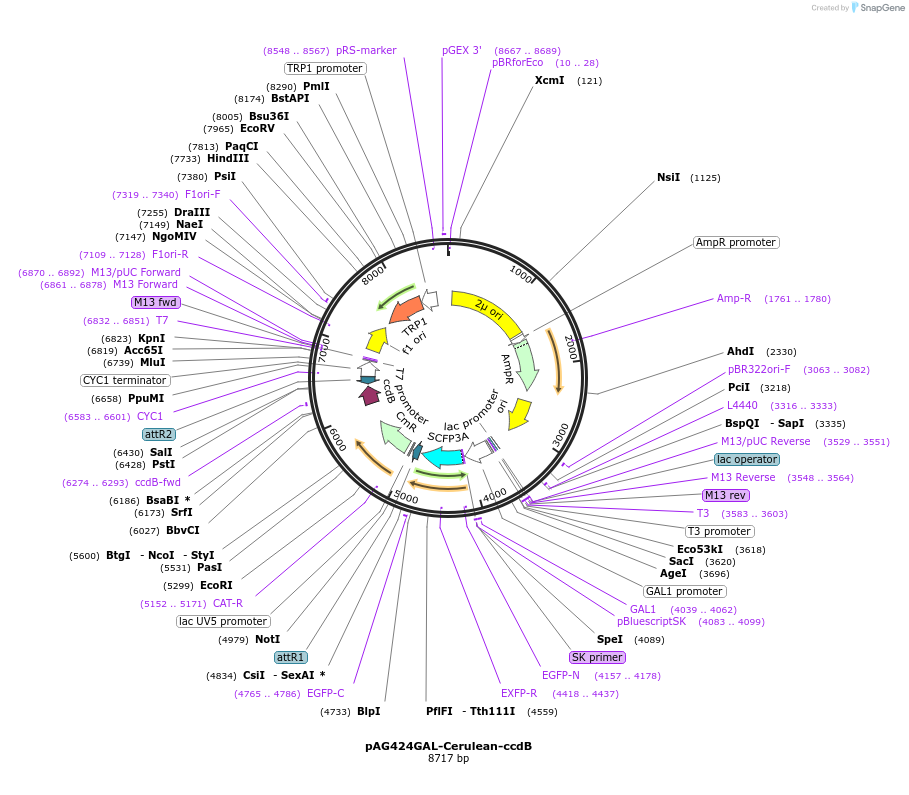
pAG424GAL-Cerulean-ccdB
Plasmid#14415DepositorTypeEmpty backboneUseGateway destinationTagsCeruleanExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
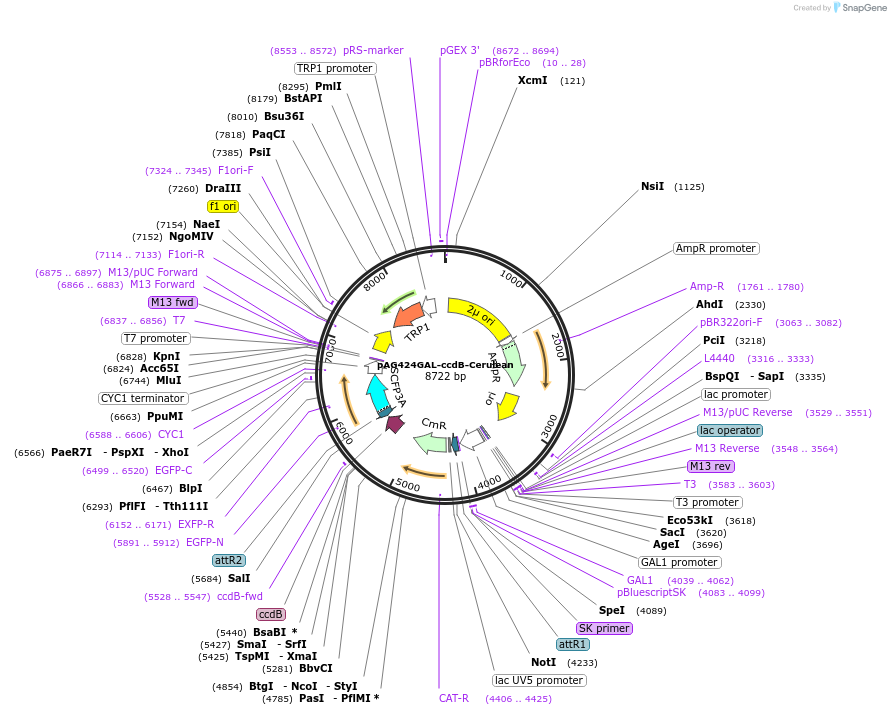
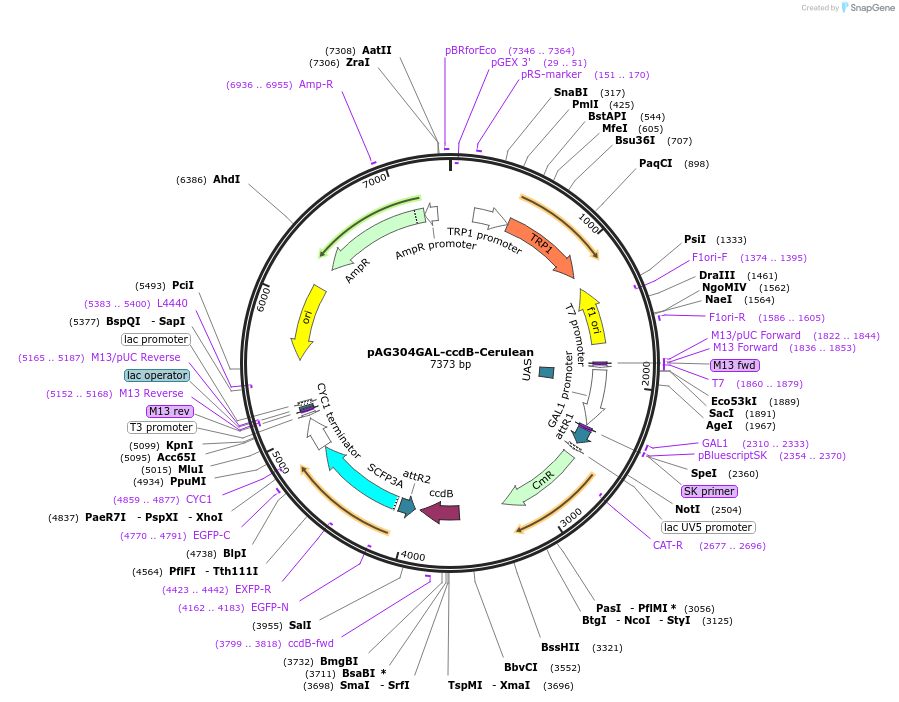
pAG424GAL-ccdB-Cerulean
Plasmid#14391DepositorTypeEmpty backboneUseGateway destinationTagsCeruleanExpressionYeastAvailable SinceMarch 29, 2007AvailabilityAcademic Institutions and Nonprofits only -
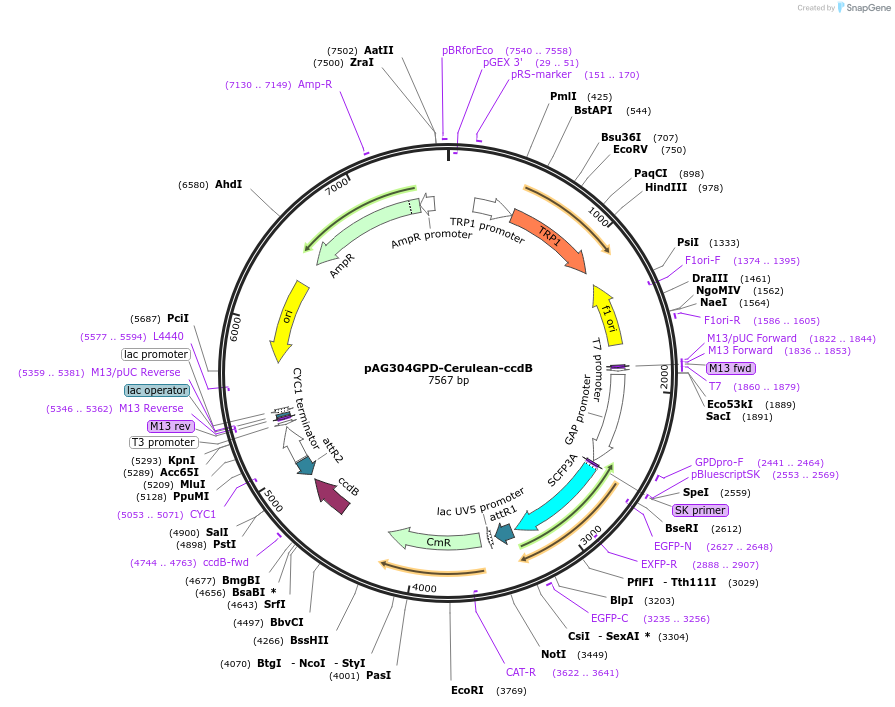
pAG304GPD-Cerulean-ccdB
Plasmid#14400DepositorTypeEmpty backboneUseGateway destinationTagsCeruleanExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
pAG304GAL-ccdB-Cerulean
Plasmid#14375DepositorTypeEmpty backboneUseGateway destinationTagsCeruleanExpressionYeastAvailable SinceMarch 29, 2007AvailabilityAcademic Institutions and Nonprofits only -
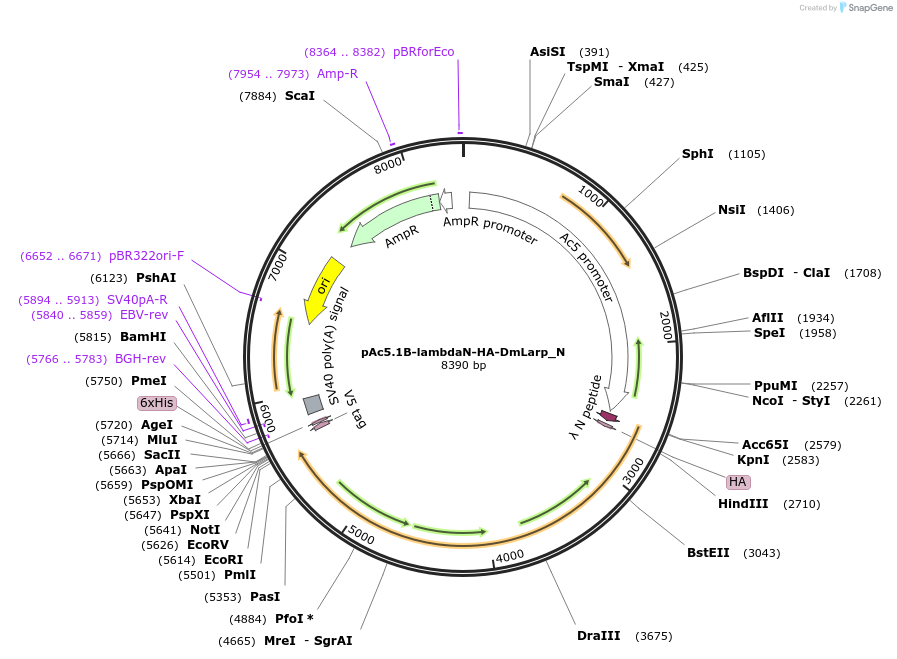
pAc5.1B-lambdaN-HA-DmLarp_N
Plasmid#146982PurposeInsect Expression of DmLarpDepositorInsertDmLarp (larp Fly)
ExpressionInsectMutationtwo non silent mutation N110T, P173L compared to …Available SinceJan. 23, 2023AvailabilityAcademic Institutions and Nonprofits only -
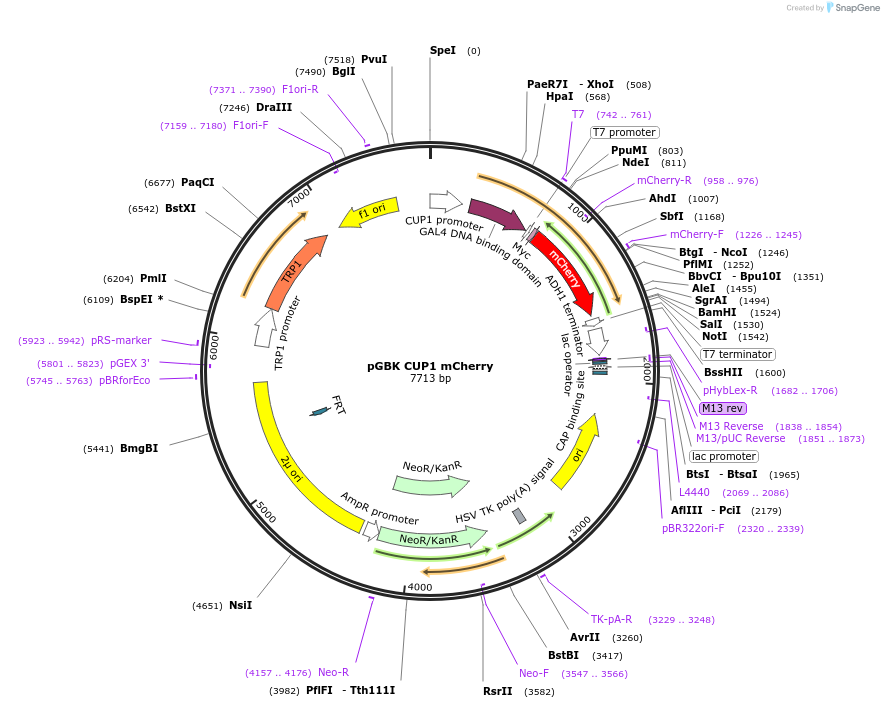
pGBK CUP1 mCherry
Plasmid#170264PurposeYeast two-hybrid vector using a copper inducible CUP1 promoter, expressing mCherryDepositorArticleInsertmomeric Cherry
TagsGAL4 DNA Binding Domain and MycExpressionBacterial and YeastPromoterCUP1Available SinceJuly 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
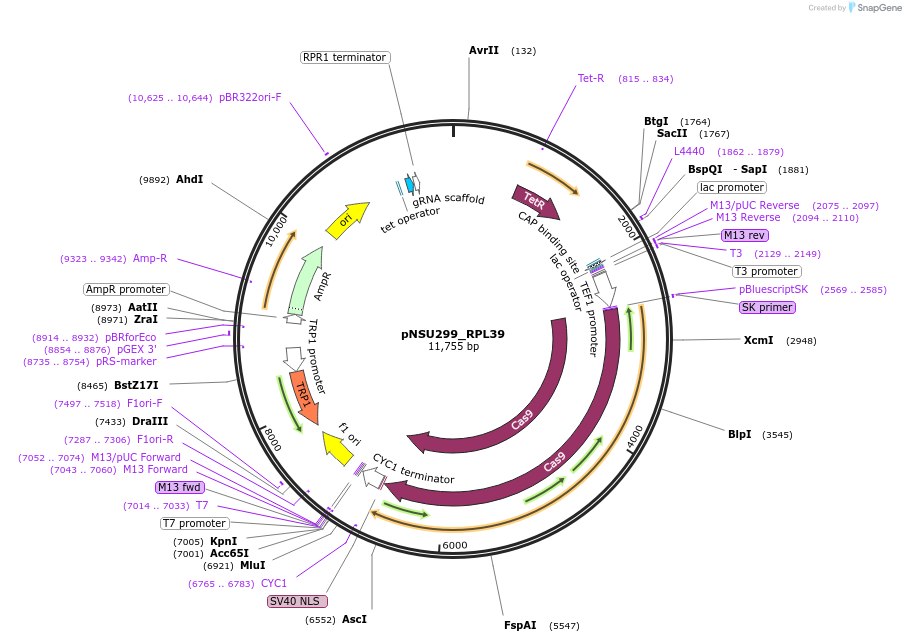
pNSU299_RPL39
Plasmid#166078PurposePlasmid for constituive spCas9 expression and tet-inducible expression of sgRNA binding to the promoter of RPL39 for double stranded break formation in yeast.DepositorInsertPromoter of RPL39 (Overlaps with 5'UTR and first base of gene) (RPL39 Budding Yeast)
UseCRISPRExpressionYeastPromoterTet-inducibleAvailable SinceApril 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
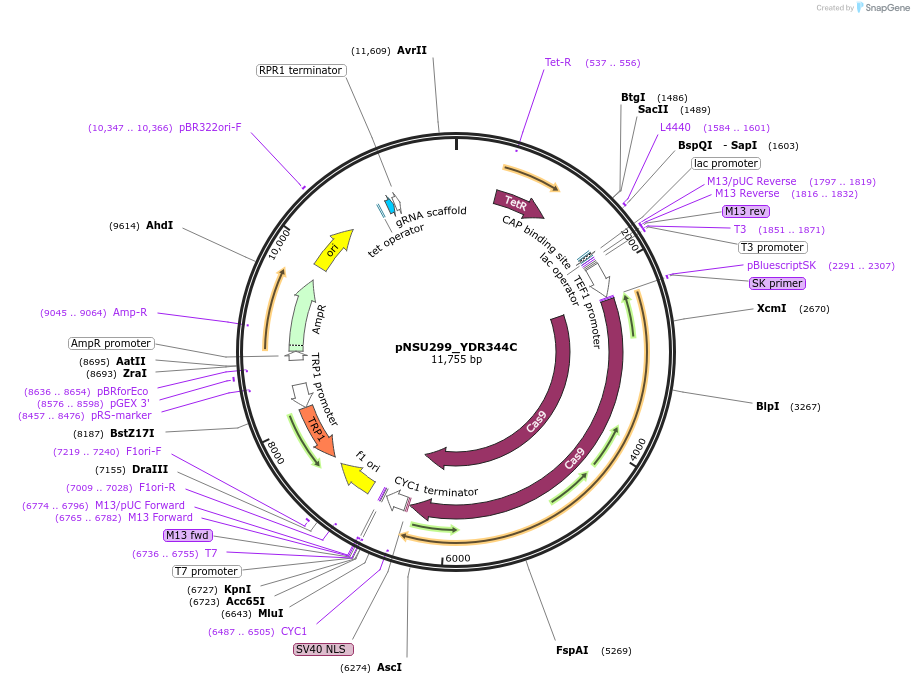
pNSU299_YDR344C
Plasmid#166072PurposePlasmid for constituive spCas9 expression and tet-inducible expression of sgRNA targeting intergenic site near HXT6 (within YDR344C) for double stranded break formation in yeast.DepositorInsertIntergenic site near HXT6 (in YDR344C, possible dubious ORF)
UseCRISPRExpressionYeastPromoterTet-inducibleAvailable SinceApril 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
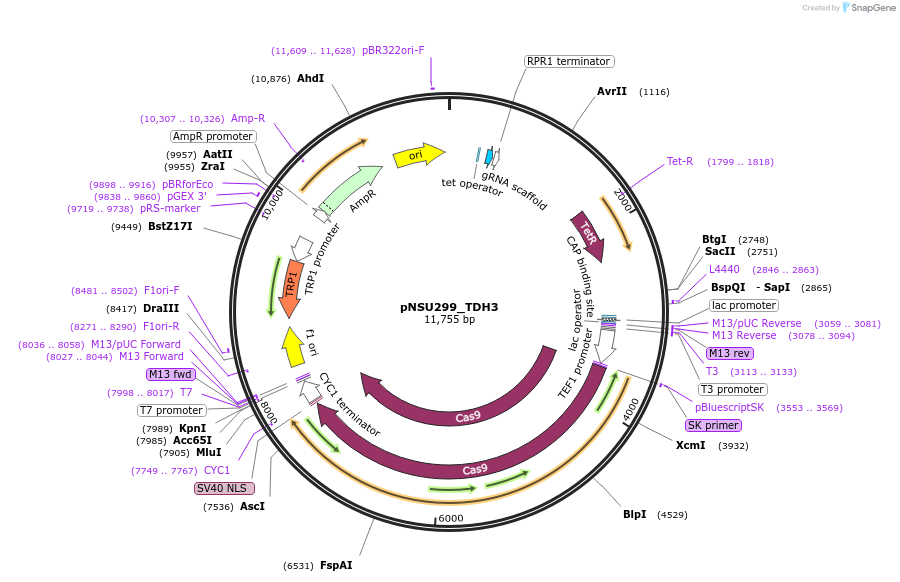
pNSU299_TDH3
Plasmid#166084PurposePlasmid for constituive spCas9 expression and tet-inducible expression of sgRNA binding to the promoter of TDH3 for double stranded break formation in yeast.DepositorAvailable SinceMarch 31, 2021AvailabilityAcademic Institutions and Nonprofits only -
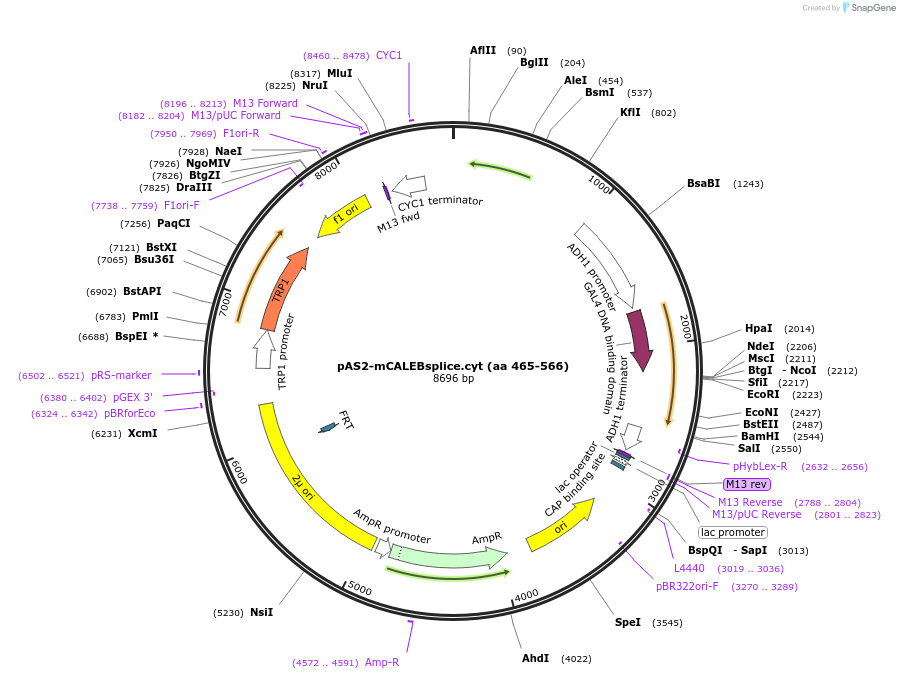
pAS2-mCALEBsplice.cyt (aa 465-566)
Plasmid#89410PurposeTwo-hybrid systemDepositorAvailable SinceMay 2, 2017AvailabilityAcademic Institutions and Nonprofits only -
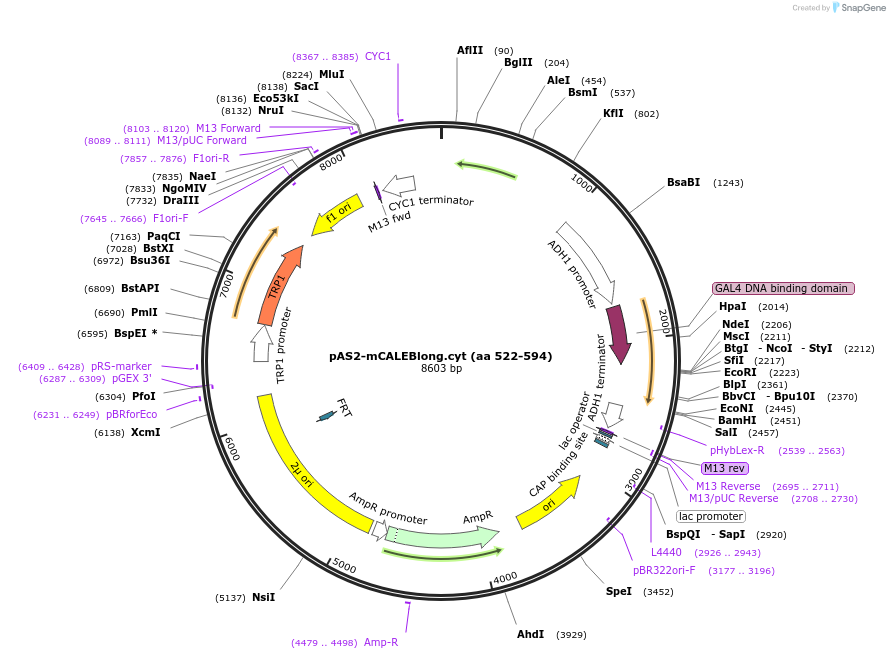
pAS2-mCALEBlong.cyt (aa 522-594)
Plasmid#89409PurposeTwo-hybrid systemDepositorAvailable SinceMay 2, 2017AvailabilityAcademic Institutions and Nonprofits only -
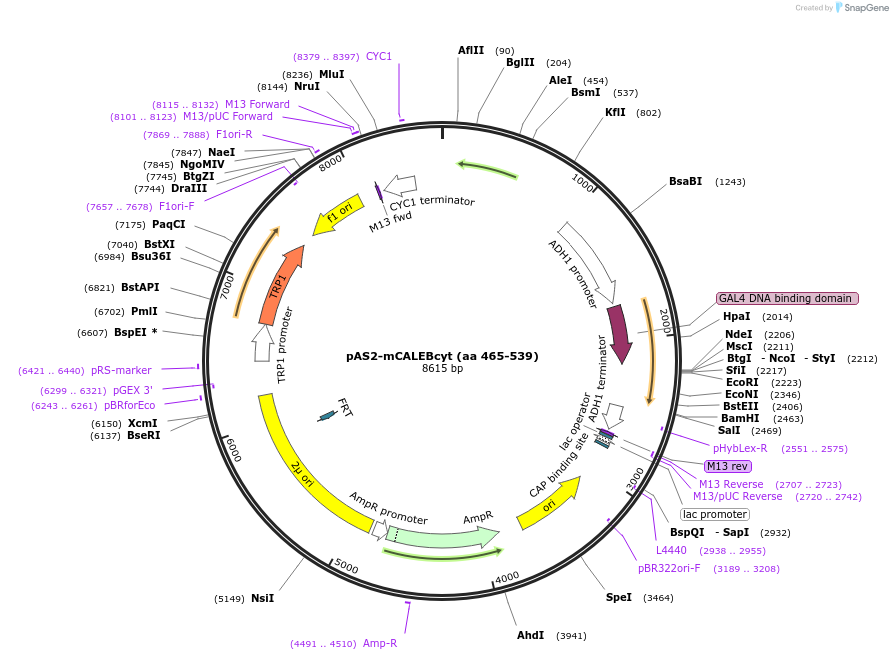
pAS2-mCALEBcyt (aa 465-539)
Plasmid#89408PurposeTwo-hybrid systemDepositorAvailable SinceMay 2, 2017AvailabilityAcademic Institutions and Nonprofits only -
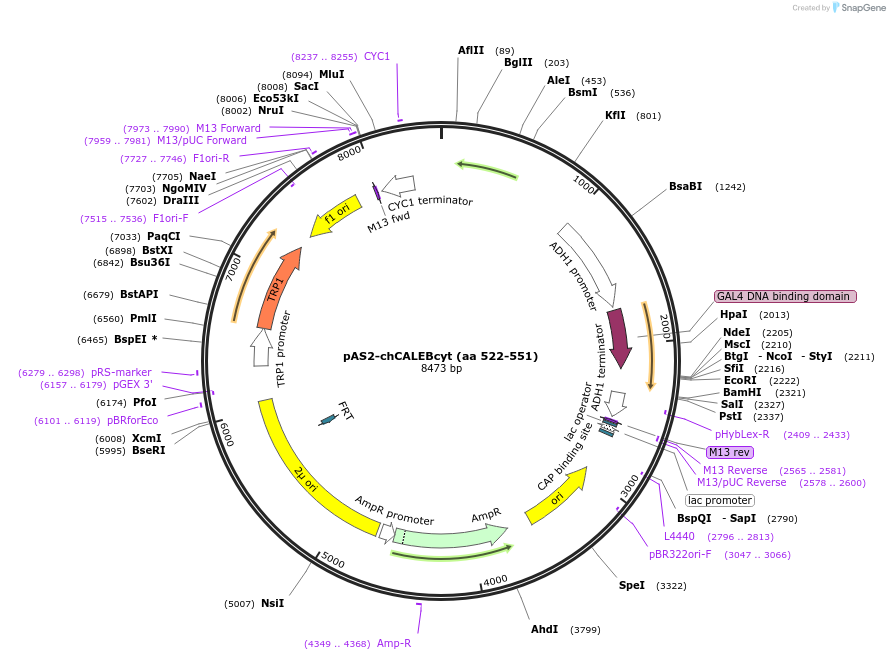
pAS2-chCALEBcyt (aa 522-551)
Plasmid#89407PurposeTwo-hybrid systemDepositorAvailable SinceMay 2, 2017AvailabilityAcademic Institutions and Nonprofits only -
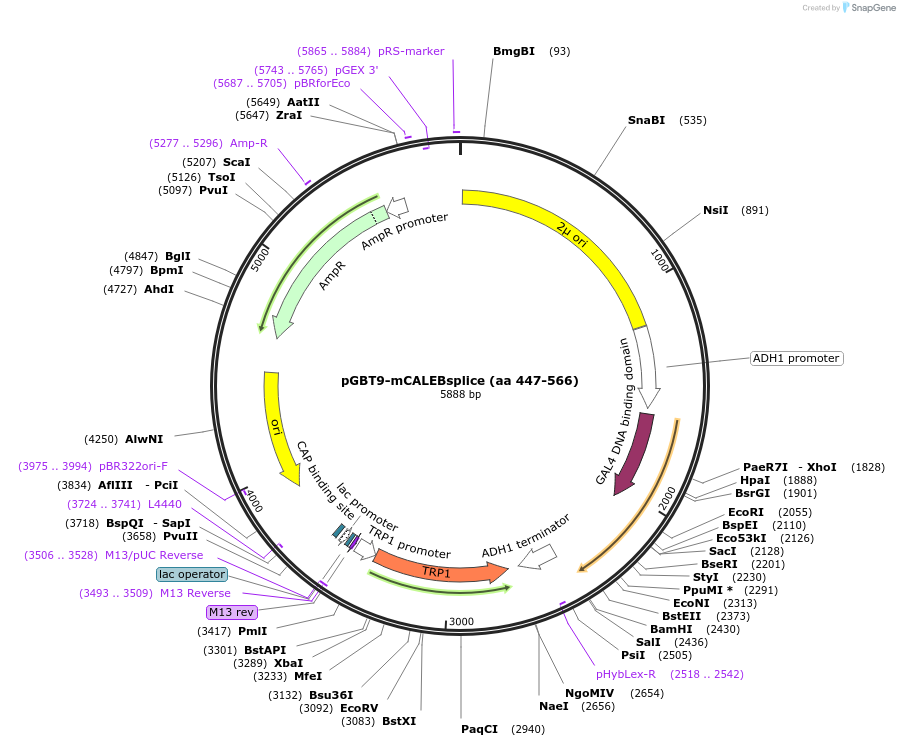
pGBT9-mCALEBsplice (aa 447-566)
Plasmid#89405PurposeTwo-hybrid systemDepositorAvailable SinceMay 2, 2017AvailabilityAcademic Institutions and Nonprofits only -
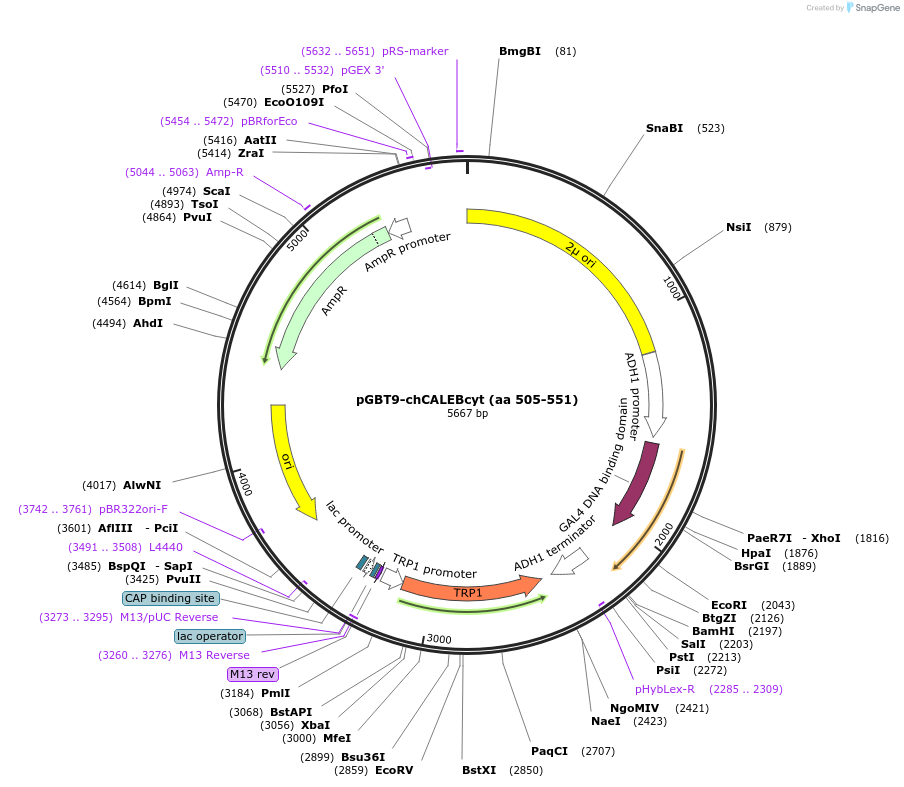
pGBT9-chCALEBcyt (aa 505-551)
Plasmid#89403PurposeTwo-hybrid systemDepositorAvailable SinceMay 2, 2017AvailabilityAcademic Institutions and Nonprofits only -
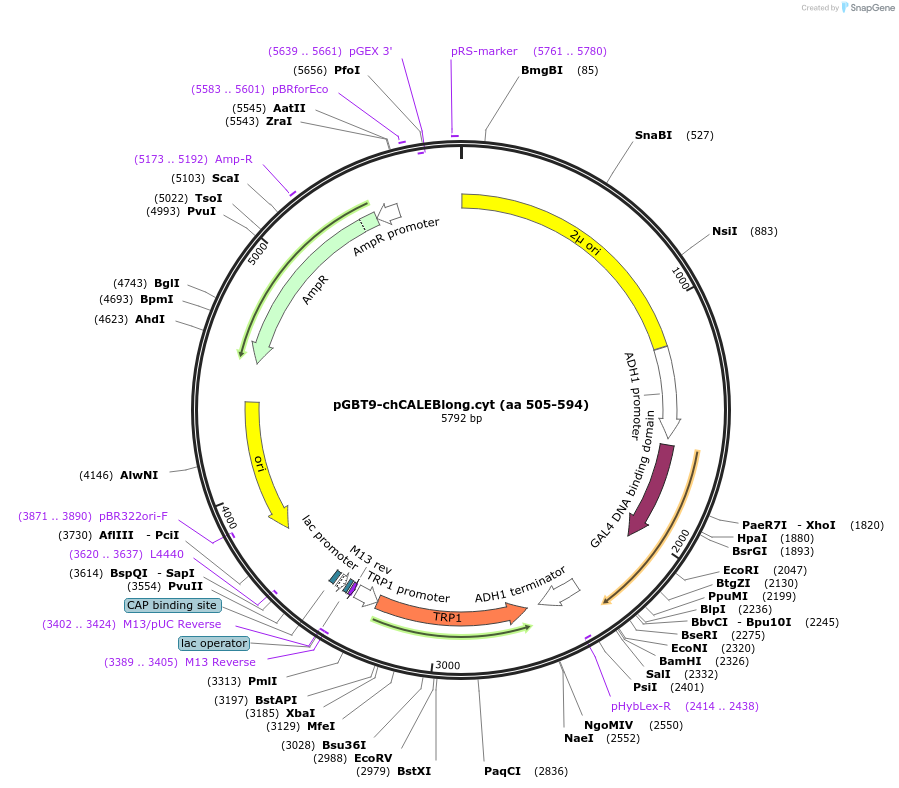
pGBT9-chCALEBlong.cyt (aa 505-594)
Plasmid#89402PurposeTwo-hybrid systemDepositorAvailable SinceMay 2, 2017AvailabilityAcademic Institutions and Nonprofits only -
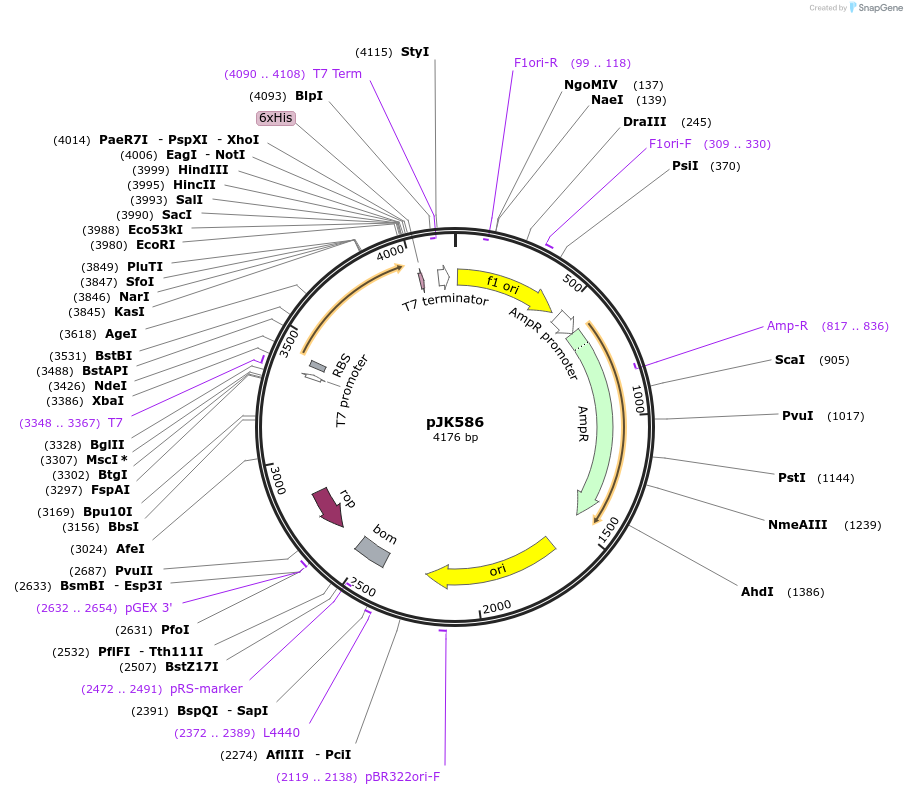
pJK586
Plasmid#72459PurposeProduces Acetobacter aceti 1023 N5-carboxyaminoimidazole ribonucleotide mutase, Trp34>Phe/Ser57>Thr/Trp165>Phe mutant (AaPurE1fwf-S57T)DepositorInsertN5-carboxyaminoimidazole ribonucleotide mutase
ExpressionBacterialMutationMutation changes tryptophan-34 to phenylalanine, …PromoterT7Available SinceFeb. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
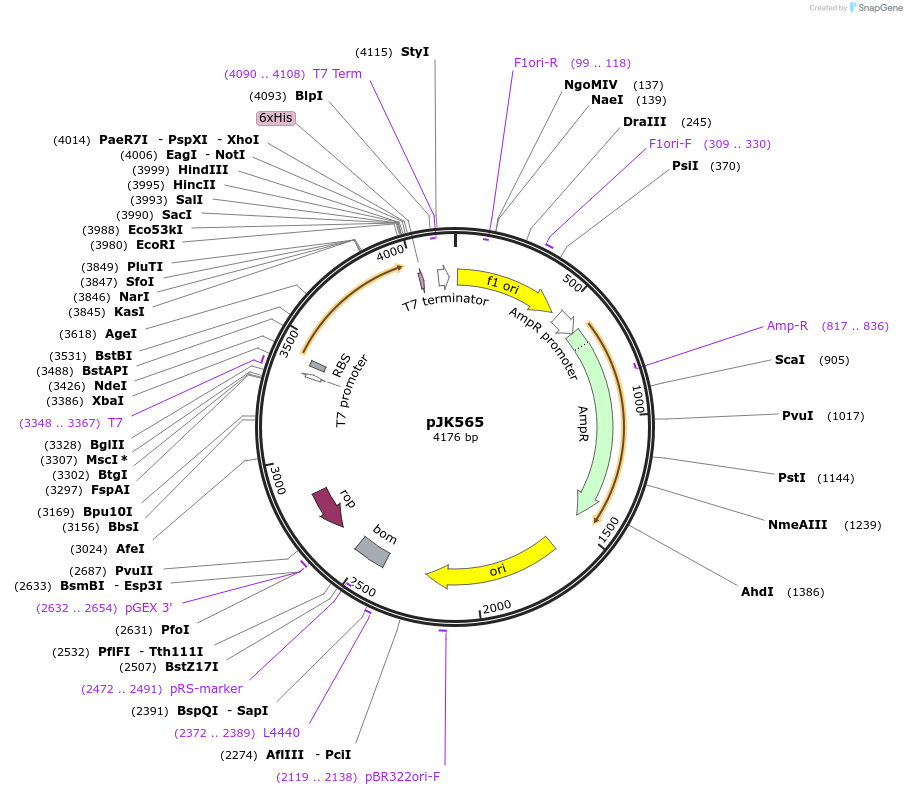
pJK565
Plasmid#72454PurposeProduces Acetobacter aceti 1023 N5-carboxyaminoimidazole ribonucleotide mutase, Trp34>Phe/His59>Ala/Trp165>Phe mutant (AaPurE1fwf-H59A)DepositorInsertN5-carboxyaminoimidazole ribonucleotide mutase
ExpressionBacterialMutationMutation changes tryptophan-34 to phenylalanine, …PromoterT7Available SinceFeb. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
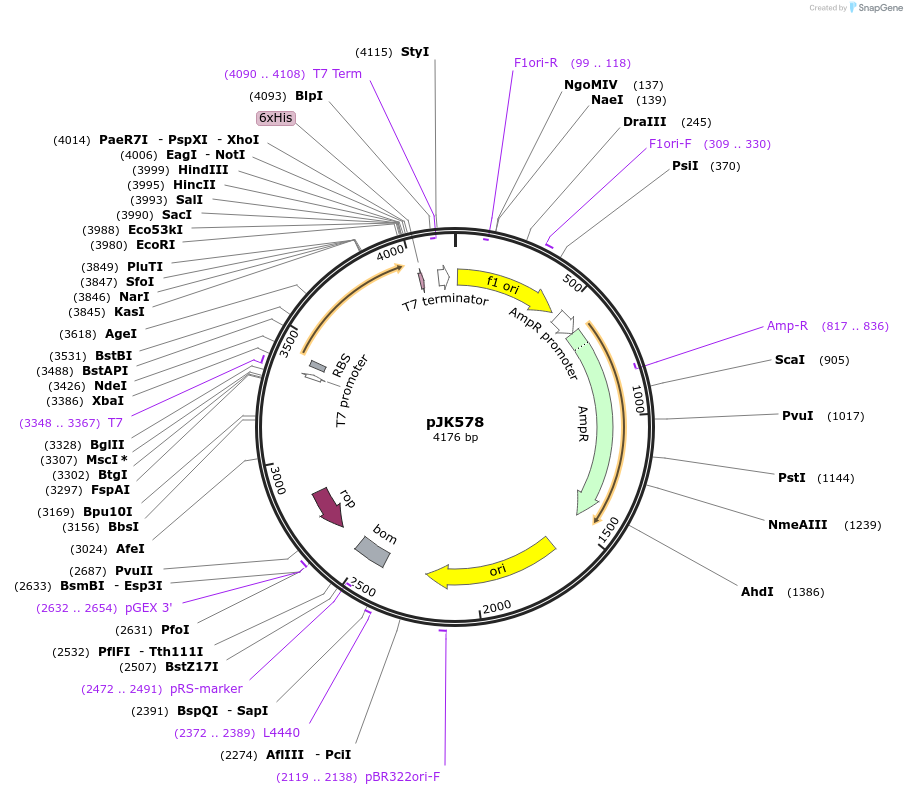
pJK578
Plasmid#72457PurposeProduces Acetobacter aceti 1023 N5-carboxyaminoimidazole ribonucleotide mutase, Trp34>Phe/Ser57>Ala/Trp165>Phe mutant (AaPurE1fwf-S57A)DepositorInsertN5-carboxyaminoimidazole ribonucleotide mutase
ExpressionBacterialMutationMutation changes tryptophan-34 to phenylalanine, …PromoterT7Available SinceFeb. 5, 2016AvailabilityAcademic Institutions and Nonprofits only -
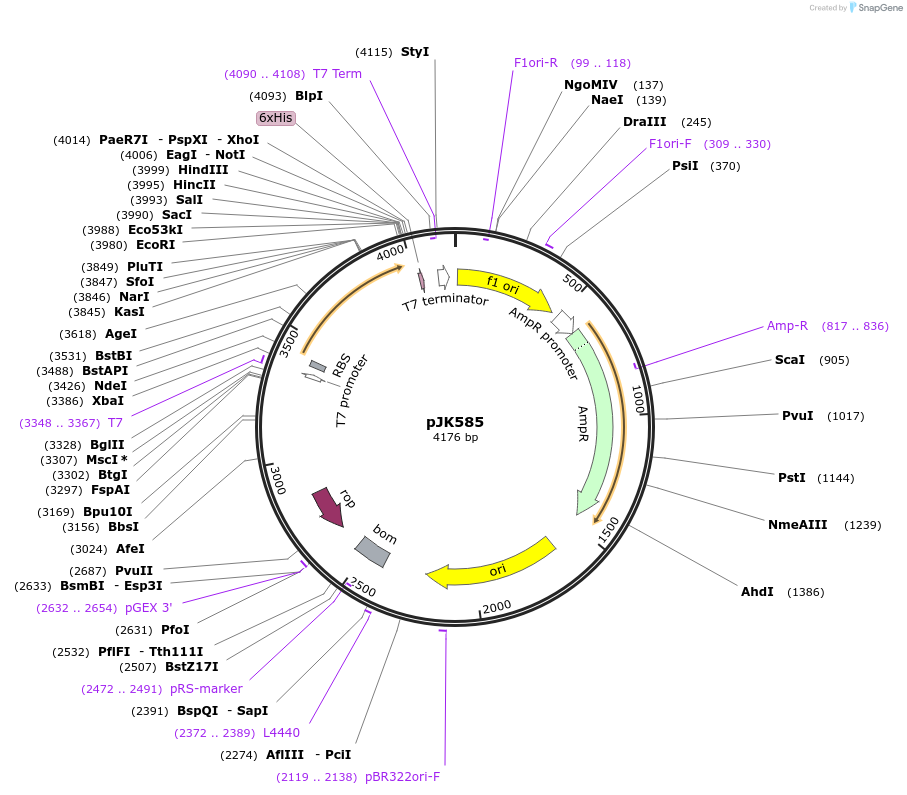
pJK585
Plasmid#72458PurposeProduces Acetobacter aceti 1023 N5-carboxyaminoimidazole ribonucleotide mutase, Trp34>Phe/Ser57>Cys/Trp165>Phe mutant (AaPurE1fwf-S57C)DepositorInsertN5-carboxyaminoimidazole ribonucleotide mutase
ExpressionBacterialMutationMutation changes tryptophan-34 to phenylalanine, …PromoterT7Available SinceJan. 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
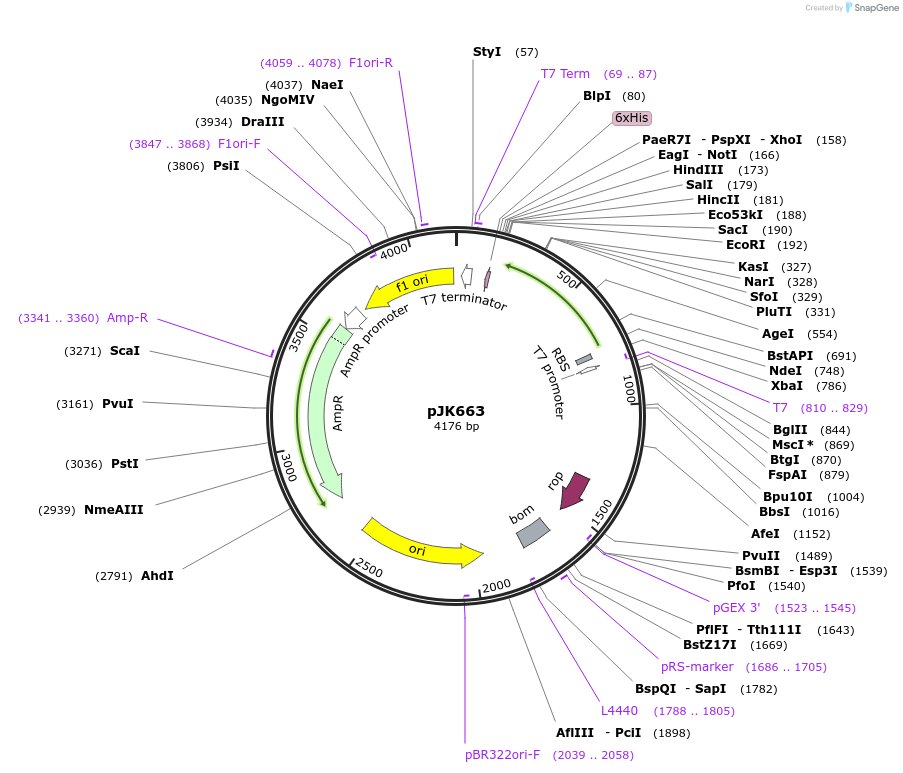
pJK663
Plasmid#71612PurposeProduces Acetobacter aceti 1023 N5-carboxyaminoimidazole ribonucleotide mutase, Tyr154>Phe/Trp165>Phe mutant (AaPurE1wwf-Y154F)DepositorInsertN5-carboxyaminoimidazole ribonucleotide mutase
ExpressionBacterialMutationchanges tyrosine-154 to phenylalanine, tryptophan…PromoterT7Available SinceJan. 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
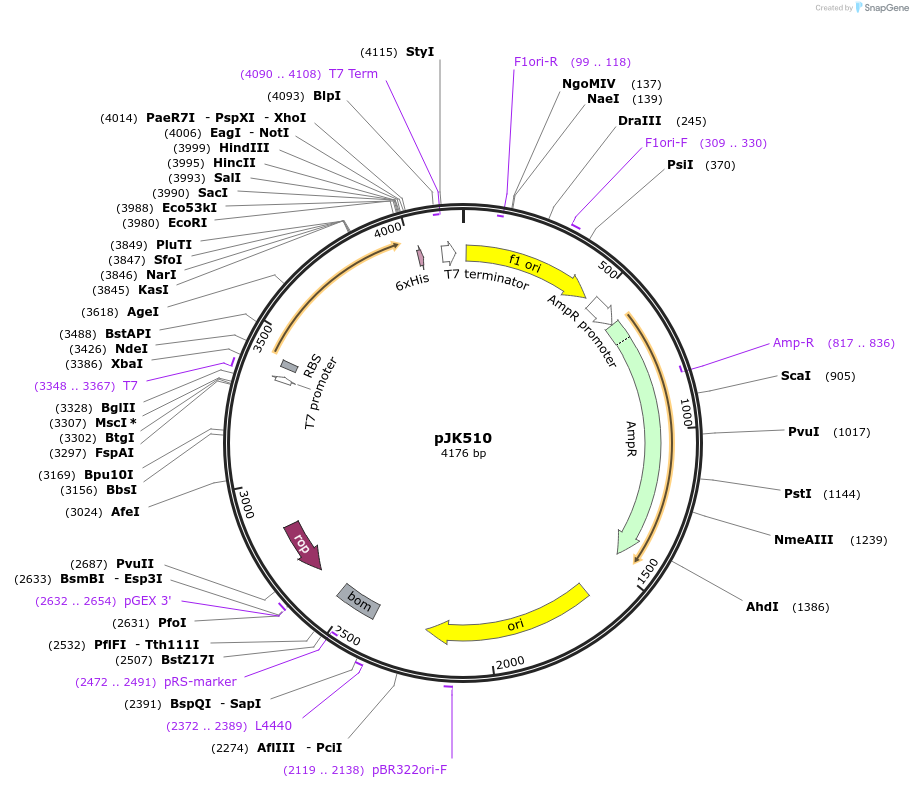
pJK510
Plasmid#71600PurposeProduces Acetobacter aceti 1023 N5-carboxyaminoimidazole ribonucleotide mutase, His59>Asn/Trp165>Phe mutant (AaPurE1wwf-H59N)DepositorInsertN5-carboxyaminoimidazole ribonucleotide mutase
ExpressionBacterialMutationchanges histidine-59 to asparagine, tryptophan-16…PromoterT7Available SinceJan. 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F22
Plasmid#23080DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed Lysine 5 to Arginine, H4 K5R.Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F24
Plasmid#23081DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed Lysine 16 to Arginine, H4 K16R.Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F25
Plasmid#23082DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed lysine 16 to Glutamine, H4 K16…Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F26
Plasmid#23083DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone 4 changed Lysine 16 to glycine, H4 K16G.Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
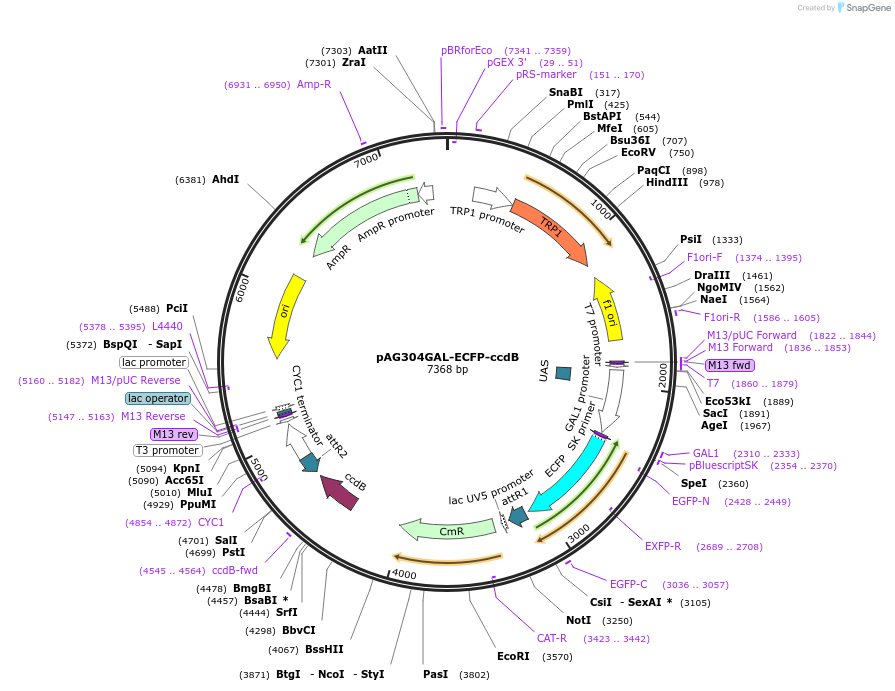
pAG304GAL-ECFP-ccdB
Plasmid#14279DepositorTypeEmpty backboneUseGateway destinationTagsECFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
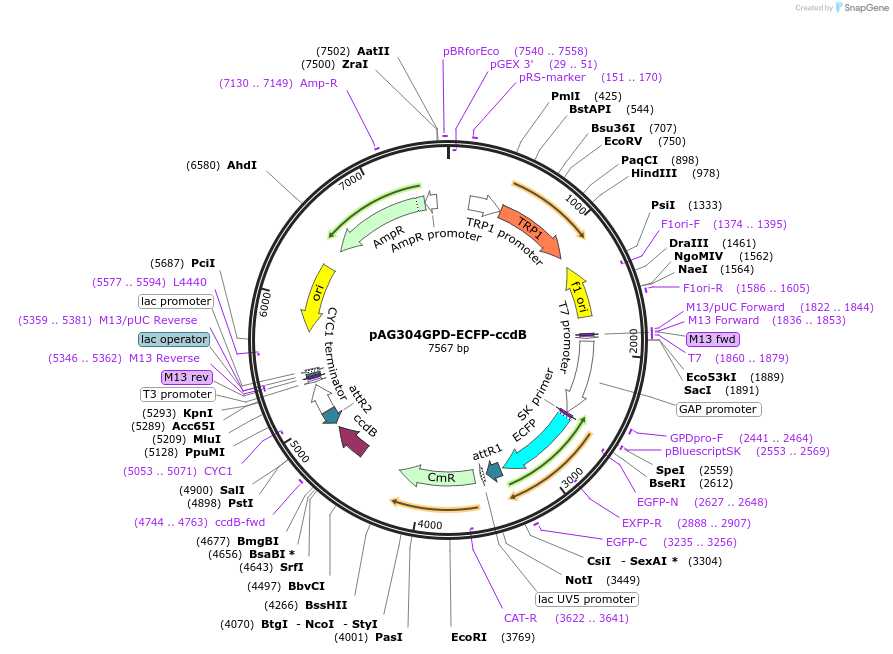
pAG304GPD-ECFP-ccdB
Plasmid#14280DepositorTypeEmpty backboneUseGateway destinationTagsECFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
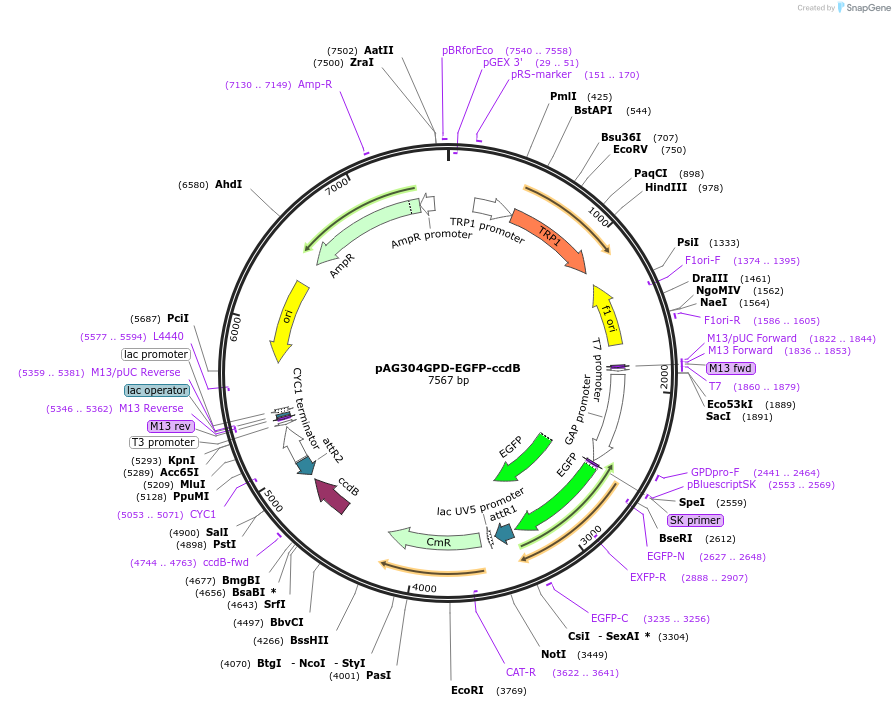
pAG304GPD-EGFP-ccdB
Plasmid#14304DepositorTypeEmpty backboneUseGateway destinationTagsEGFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only