We narrowed to 22,770 results for: ARC
-
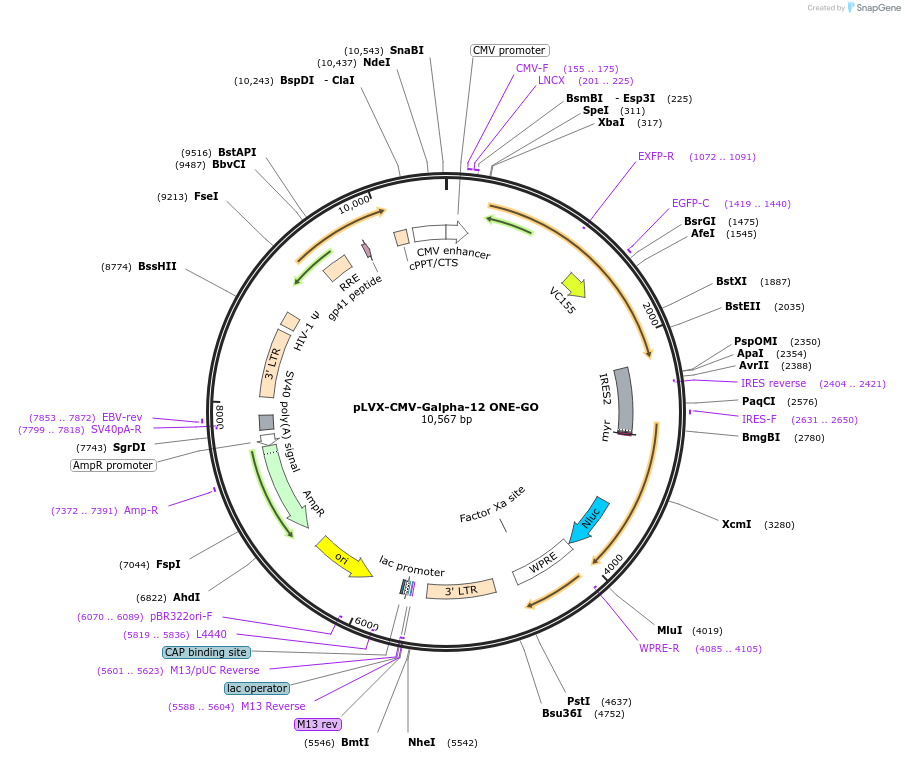
Plasmid#189739PurposeGPCR/G protein BRET biosensorDepositorInsertsUseLentiviralAvailable SinceJan. 19, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits
-
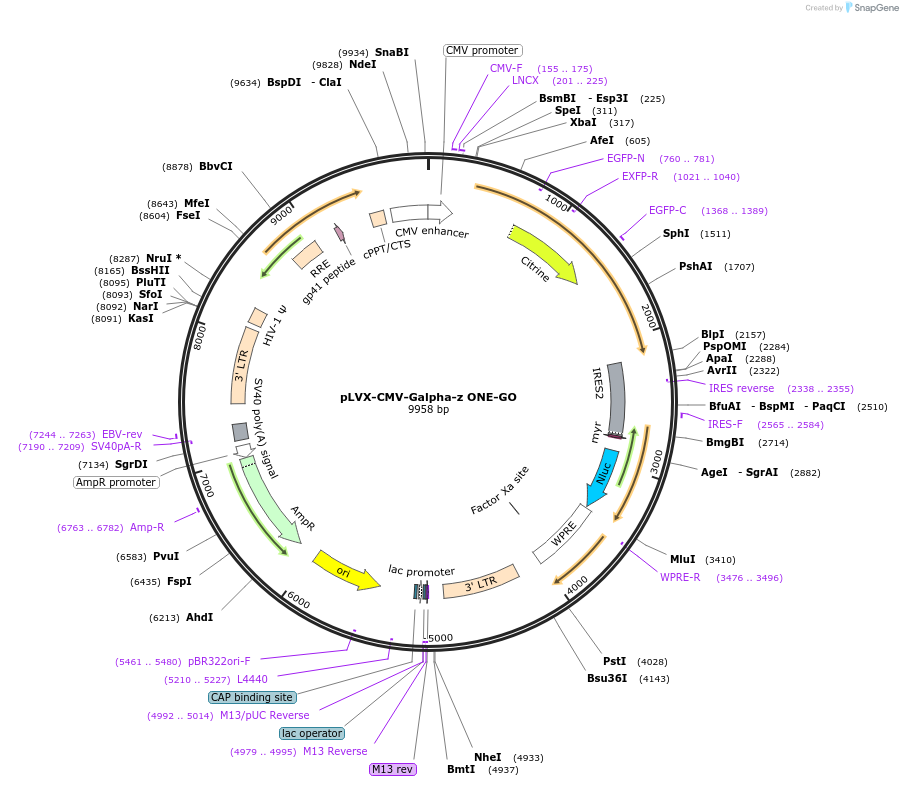
pLVX-CMV-Galpha-z ONE-GO
Plasmid#189734PurposeGPCR/G protein BRET biosensorDepositorInsertsUseLentiviralAvailable SinceJan. 19, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
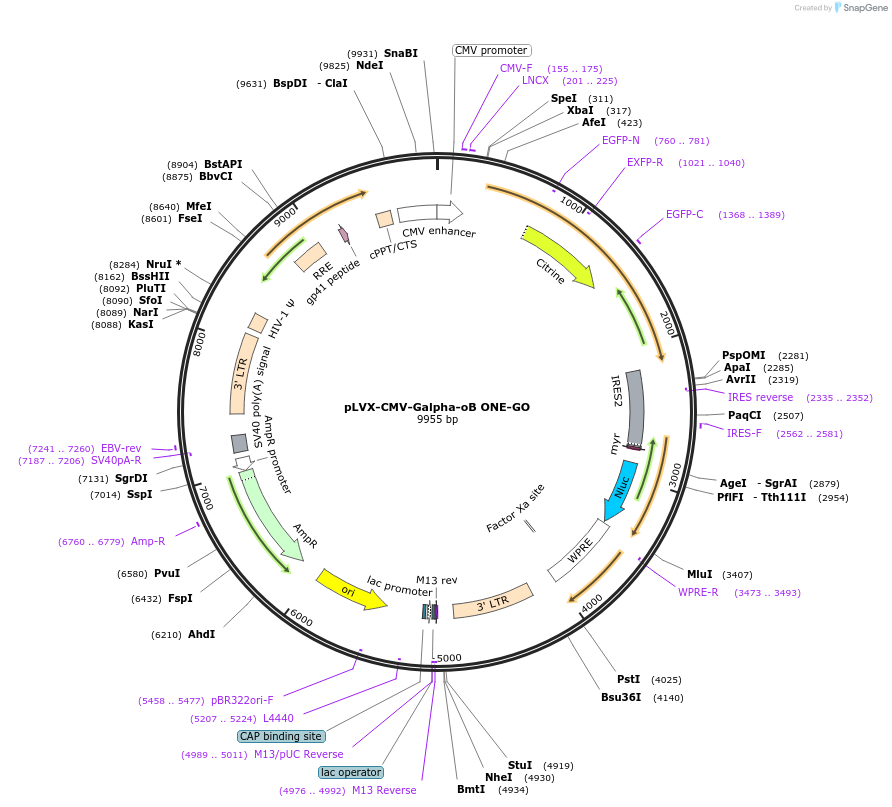
pLVX-CMV-Galpha-oB ONE-GO
Plasmid#189733PurposeGPCR/G protein BRET biosensorDepositorInsertsUseLentiviralAvailable SinceJan. 19, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
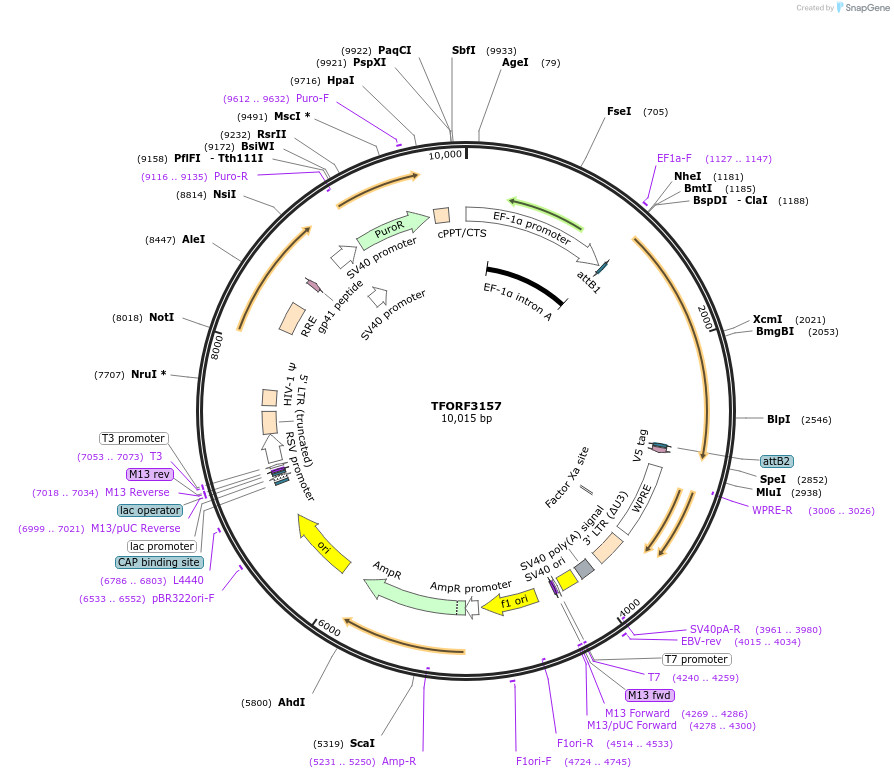
TFORF3157
Plasmid#144633PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
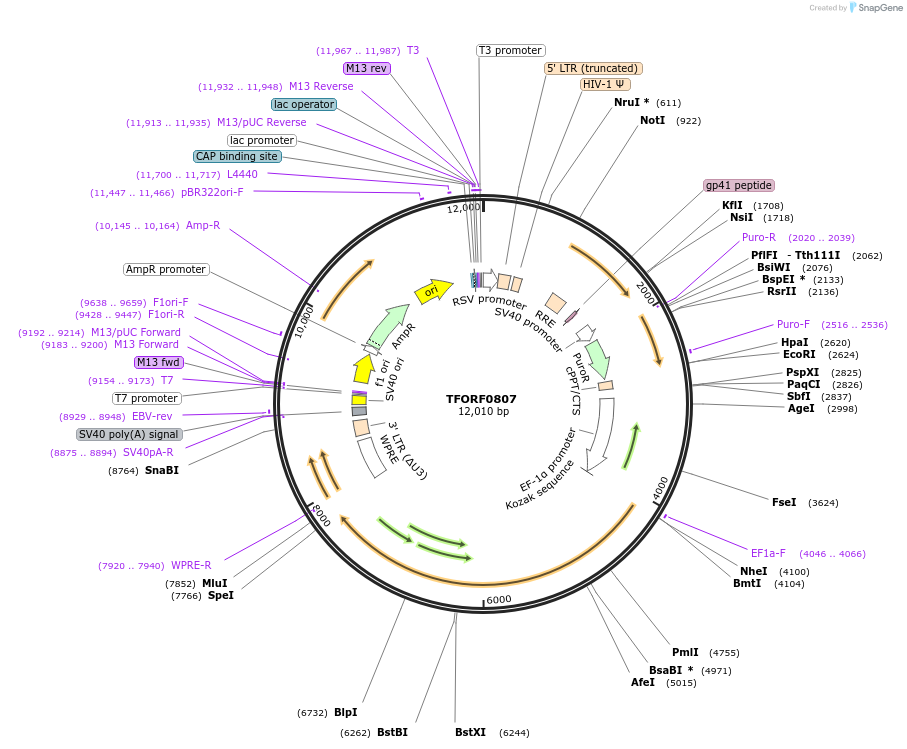
TFORF0807
Plasmid#141712PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
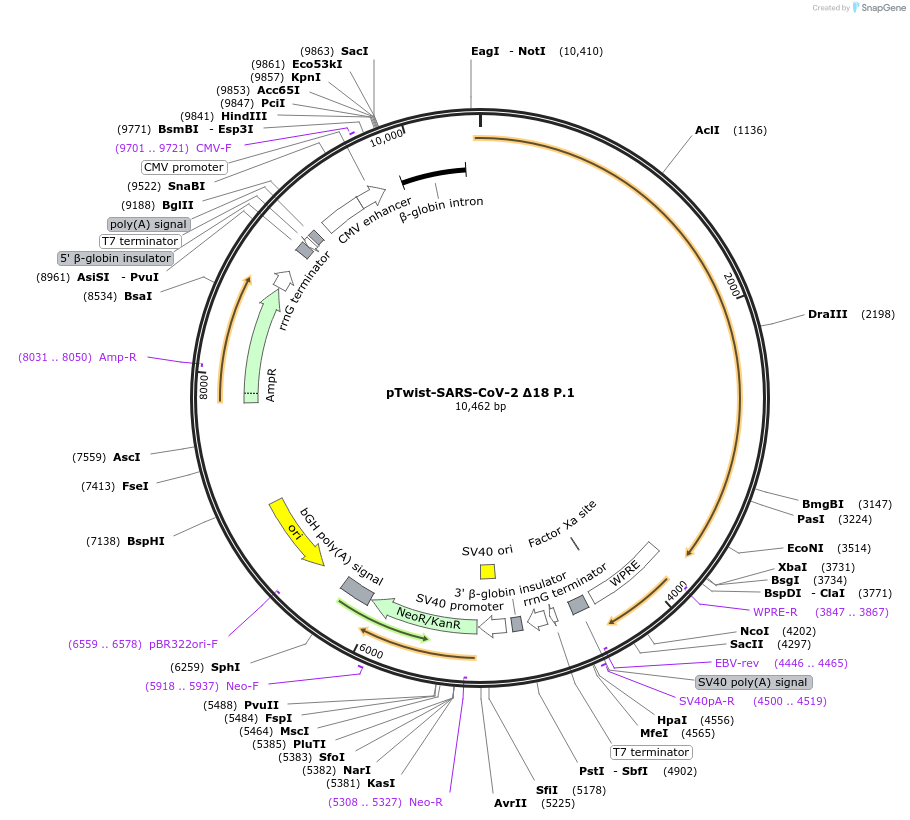
pTwist-SARS-CoV-2 Δ18 P.1
Plasmid#173476PurposeEncodes SARS-CoV-2 P.1 Spike Protein lacking 18 C-terminal amino acids for pseudovirus productionDepositorInsertSARS-CoV-2 Spike Truncated to remove 18 amino acids
ExpressionMammalianAvailable SinceAug. 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
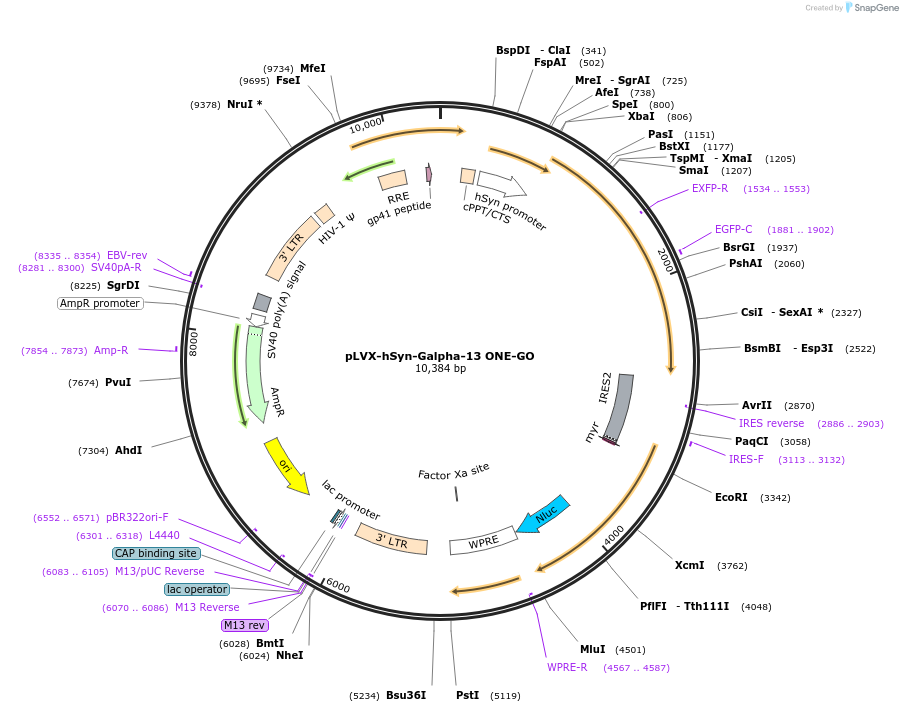
pLVX-hSyn-Galpha-13 ONE-GO
Plasmid#204337PurposeGPCR/G protein BRET biosensorDepositorInserthSyn Galpha-13 ONE-GO (GNA13 Human)
UseLentiviralAvailable SinceJan. 19, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
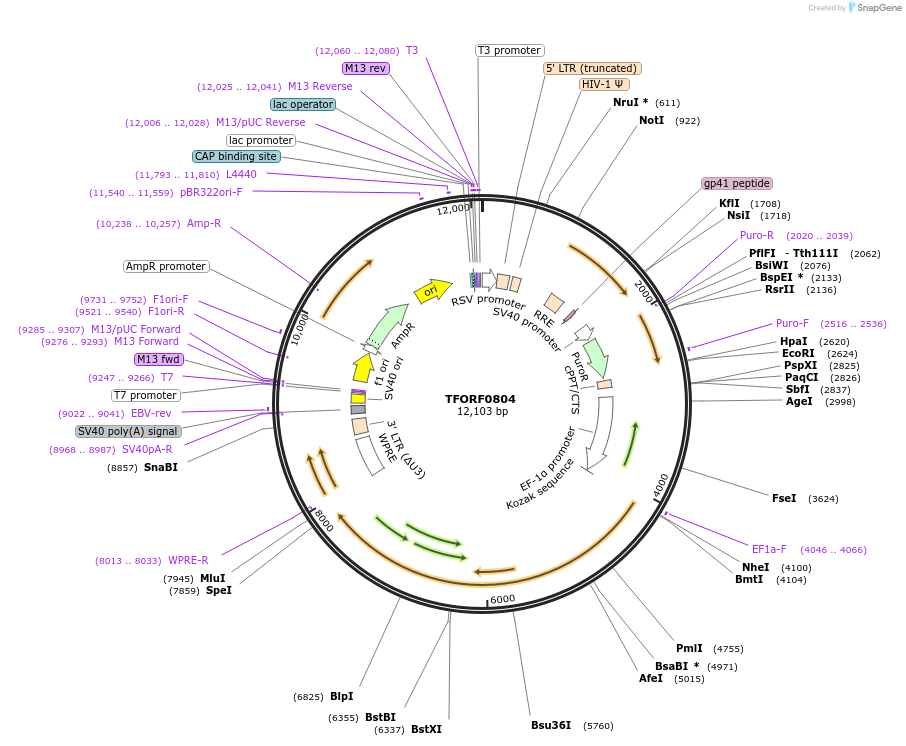
TFORF0804
Plasmid#141709PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
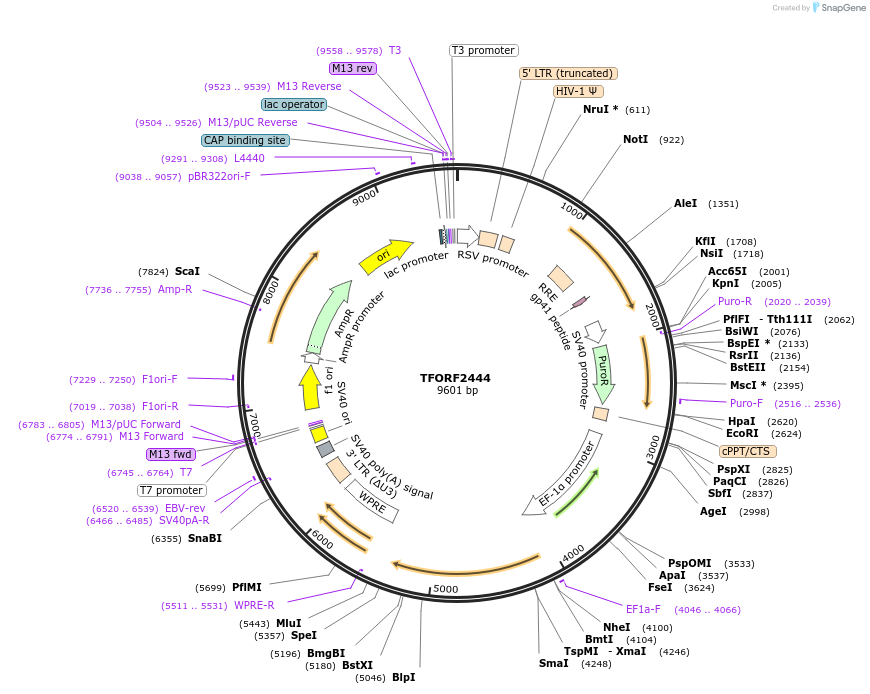
TFORF2444
Plasmid#142081PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJune 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
TFORF0805
Plasmid#141710PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
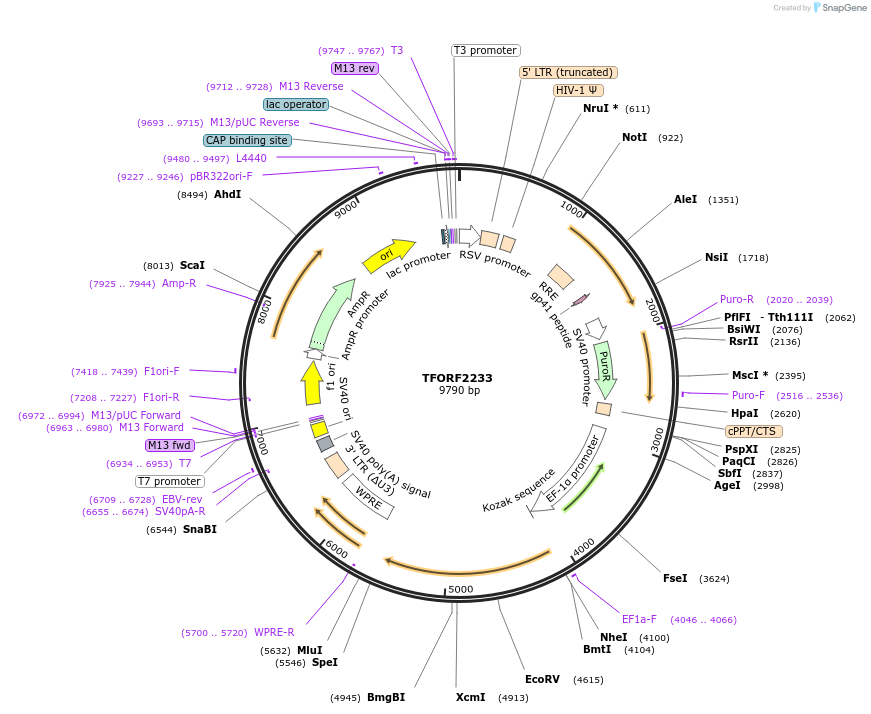
TFORF2233
Plasmid#141981PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJune 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
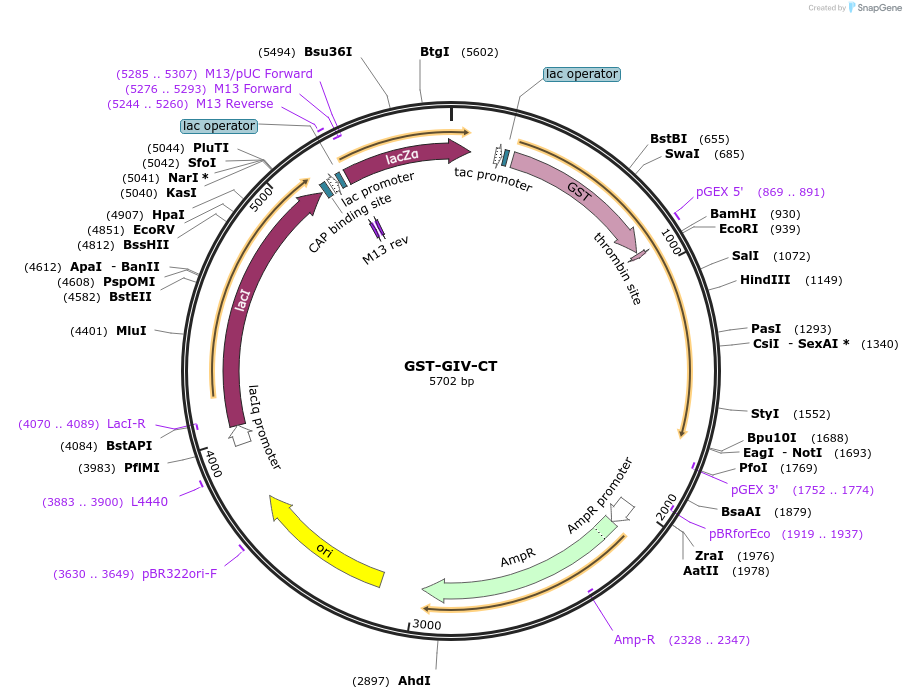
GST-GIV-CT
Plasmid#69863PurposeBacterial expression of GST tagged GIV-CTDepositorInsertGIV/Girdin (CCDC88A Human)
TagsGSTExpressionBacterialMutationC terminal amino acids 1623-1870 of human GIVPromoterTACAvailable SinceNov. 4, 2015AvailabilityAcademic Institutions and Nonprofits only -
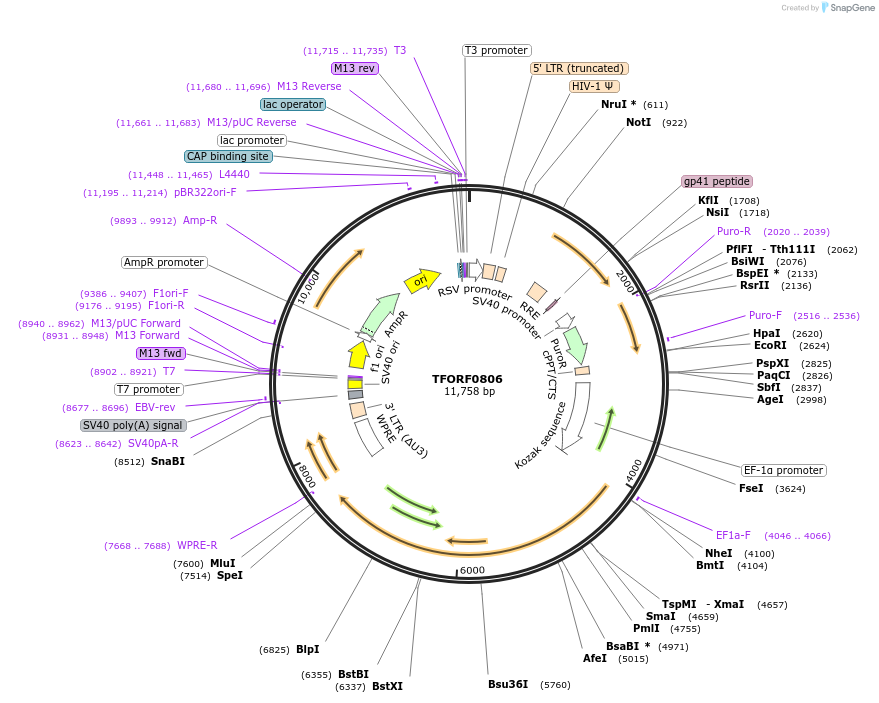
TFORF0806
Plasmid#141711PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
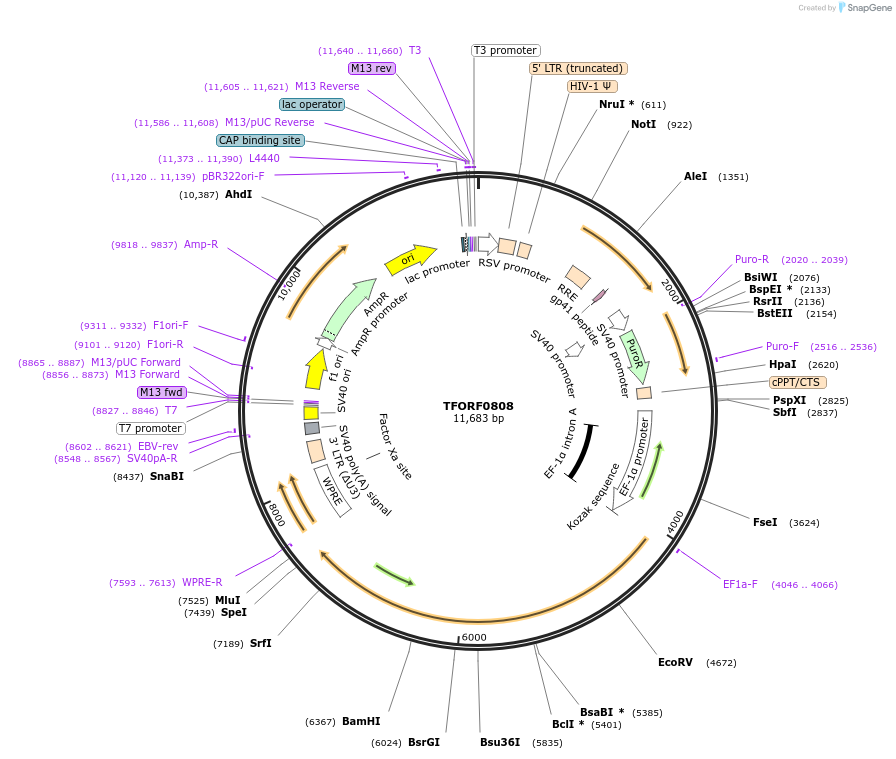
TFORF0808
Plasmid#142693PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceMay 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
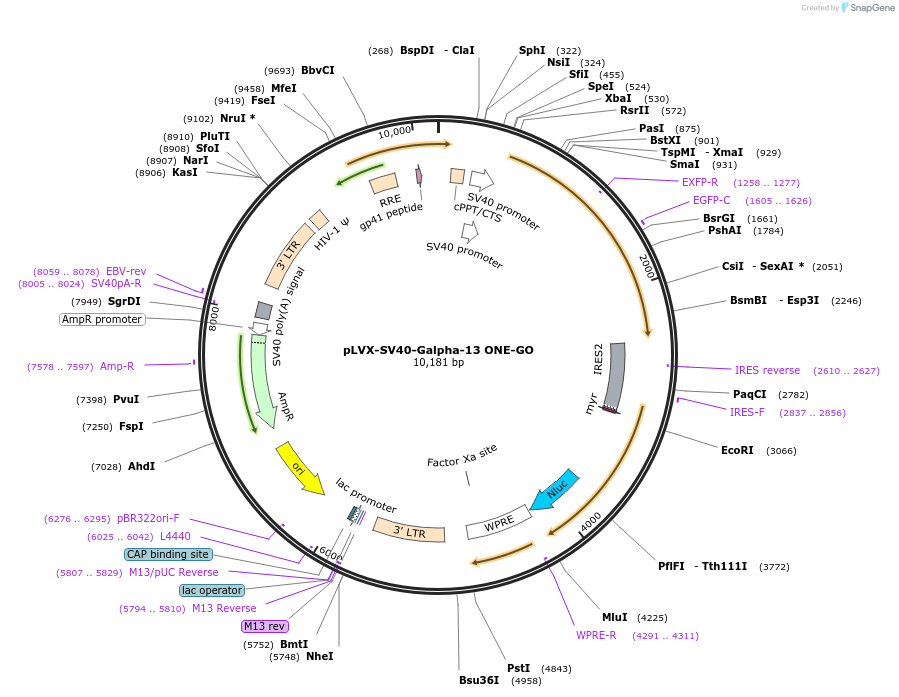
pLVX-SV40-Galpha-13 ONE-GO
Plasmid#204332PurposeGPCR/G protein BRET biosensorDepositorInsertSV40 Galpha-13 ONE-GO (GNA13 Human)
UseLentiviralAvailable SinceJan. 19, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
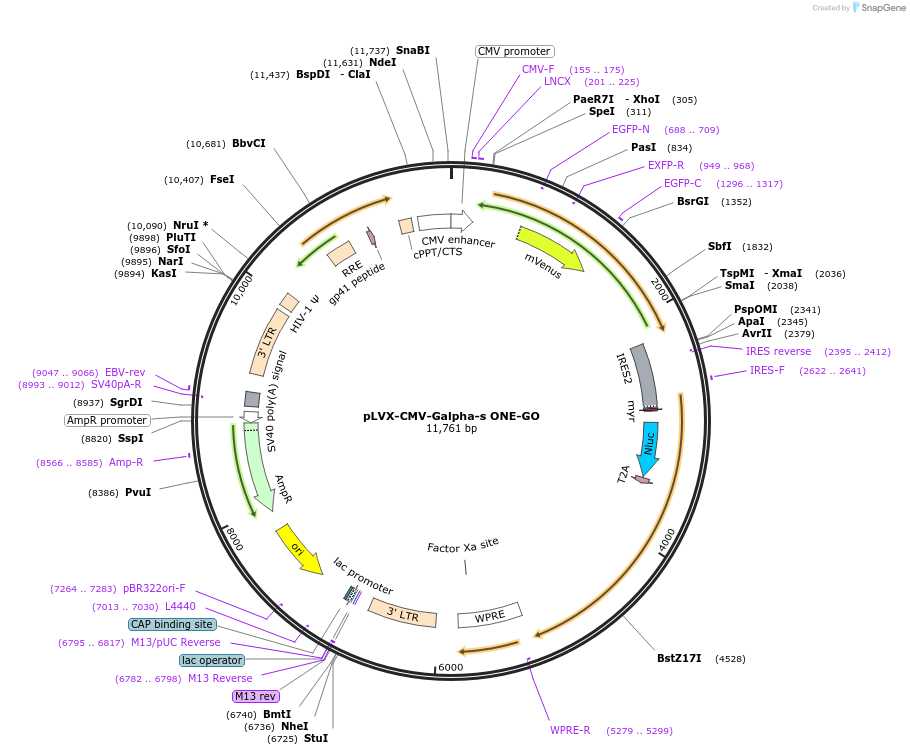
pLVX-CMV-Galpha-s ONE-GO
Plasmid#189731PurposeGPCR/G protein BRET biosensorDepositorInsertsUseLentiviralAvailable SinceJan. 19, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
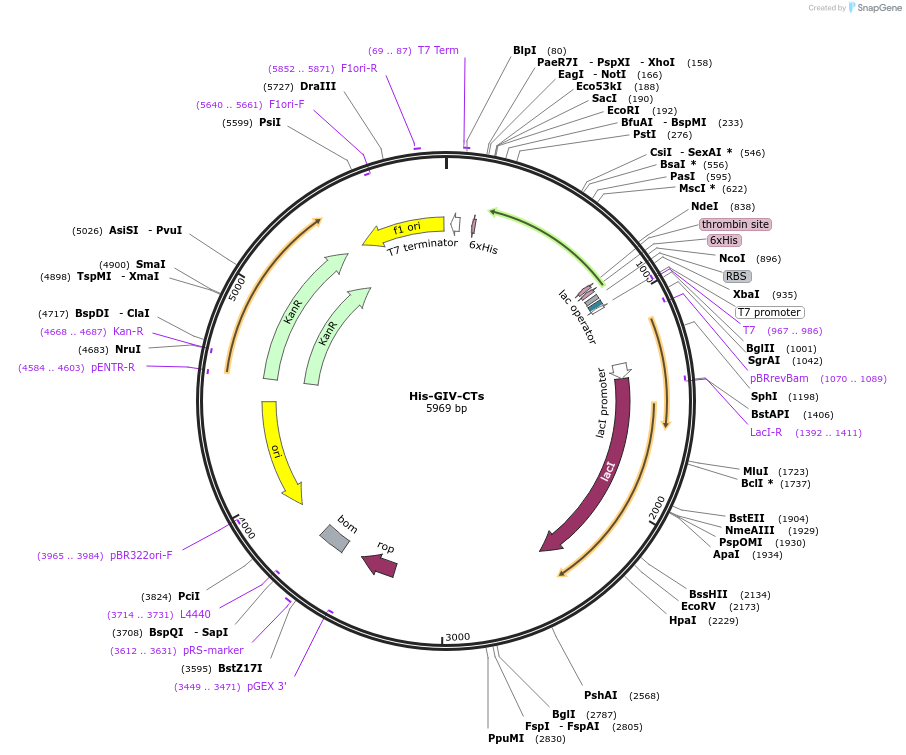
His-GIV-CTs
Plasmid#69864PurposeExpress His tagged GIV-CTs (1660–1870).DepositorInsertGIV/Girdin (CCDC88A Human)
TagsHisExpressionBacterialMutationC terminal amino acids S1660-S1870 of human GIV.PromoterT7Available SinceNov. 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
p3xFLAG-CMV14-GINIP
Plasmid#204845PurposeExpress 3x FLAG tagged-GINIP in mammalian cellsDepositorAvailable SinceSept. 14, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
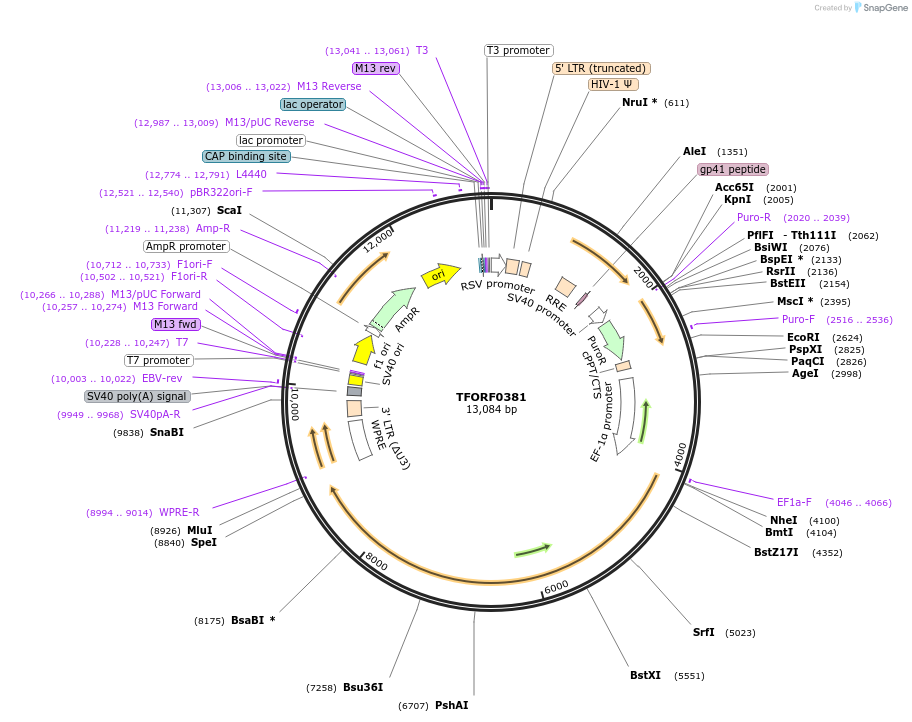
TFORF0381
Plasmid#142256PurposeLentiviral vector for overexpressing transcription factor ORFs with unique 24-bp barcodes. Barcodes facilitate identification of transcription factors in pooled screens.DepositorAvailable SinceDec. 2, 2024AvailabilityAcademic Institutions and Nonprofits only -
pTwist-SARS-CoV Δ18
Plasmid#169465PurposeEncodes SARS-CoV Spike Protein lacking 18 C-terminal amino acids for pseudovirus productionDepositorInsertSARS-CoV Spike truncated to remove 18 amino acids (S )
ExpressionMammalianAvailable SinceJune 21, 2021AvailabilityAcademic Institutions and Nonprofits only