We narrowed to 81,061 results for: myc
-
Plasmid#57832DepositorAvailable SinceOct. 24, 2014AvailabilityAcademic Institutions and Nonprofits only
-
mKO-Actin-7
Plasmid#57830DepositorAvailable SinceOct. 24, 2014AvailabilityAcademic Institutions and Nonprofits only -
mKOet-CENPB-N-22
Plasmid#57912PurposeLocalization: Nucleus/Centromeres, Excitation: 548, Emission: 561DepositorInsertCENPB (CENPB Human)
TagsmKOetExpressionMammalianMutationaa 1-169 of CENPB (DNA Binding Domain)PromoterCMVAvailable SinceOct. 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
mTurquoise-H1-10
Plasmid#55555PurposeLocalization: Nucleus/Histones, Excitation: 434, Emission: 474DepositorAvailable SinceOct. 2, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
pFX05
Plasmid#44531DepositorInsertHHT2
ExpressionYeastMutationK56QAvailable SinceMay 23, 2013AvailabilityAcademic Institutions and Nonprofits only -
pCM350
Plasmid#44549DepositorInsertHA-htz1
TagsHAExpressionYeastMutationdeleted amino acids 1-22Available SinceMay 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
pFX06
Plasmid#44532DepositorInsertHHT2
ExpressionYeastMutationK56RAvailable SinceMay 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
pTH672-SUP35-Y351C
Plasmid#29733DepositorInsertSUP35 gene encoding translation release factor 3 from S. cerevisiae (SUP35 Budding Yeast)
ExpressionYeastMutationY351C mutant gene of SUP35 including genomic sequ…Available SinceSept. 6, 2011AvailabilityAcademic Institutions and Nonprofits only -
FC511
Plasmid#27330DepositorAvailable SinceFeb. 28, 2011AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F22
Plasmid#23080DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed Lysine 5 to Arginine, H4 K5R.Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F24
Plasmid#23081DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed Lysine 16 to Arginine, H4 K16R.Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F25
Plasmid#23082DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed lysine 16 to Glutamine, H4 K16…Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F26
Plasmid#23083DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone 4 changed Lysine 16 to glycine, H4 K16G.Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
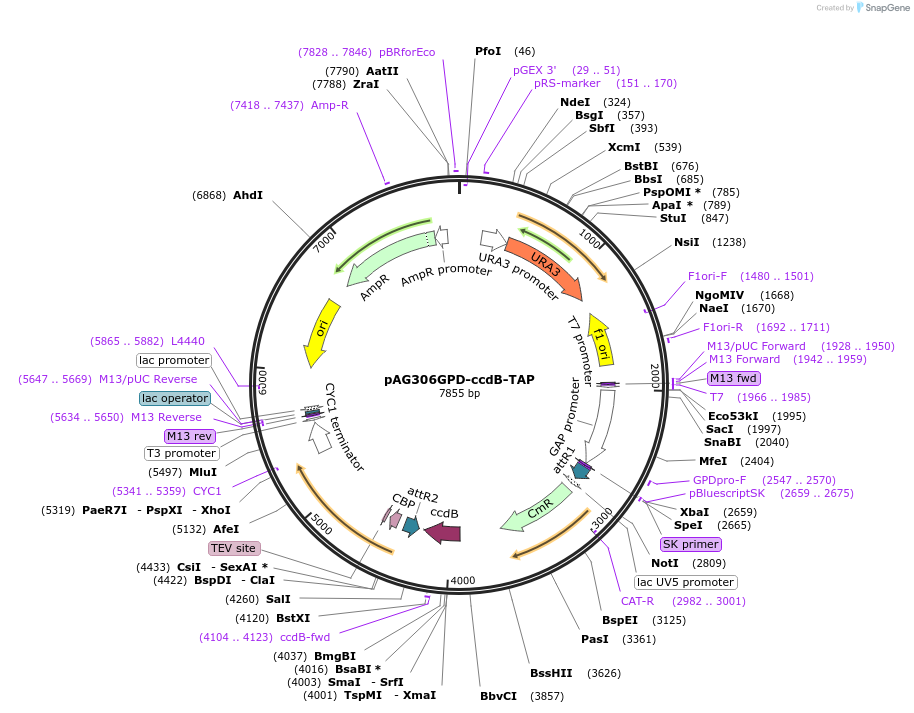
pAG306GPD-ccdB-TAP
Plasmid#14260DepositorTypeEmpty backboneUseGateway destinationTagsTAPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
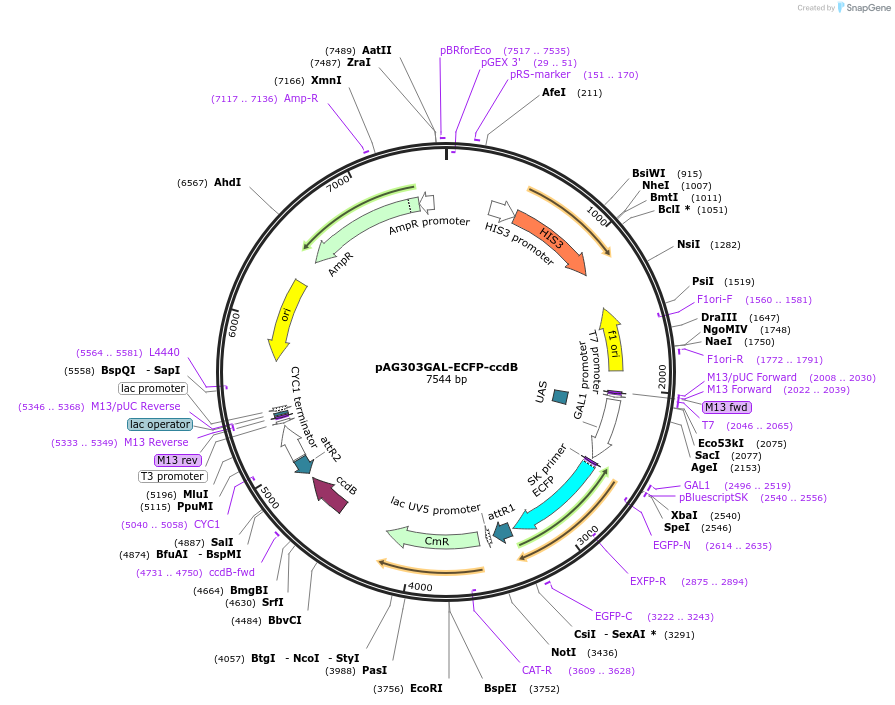
pAG303GAL-ECFP-ccdB
Plasmid#14277DepositorTypeEmpty backboneUseGateway destinationTagsECFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
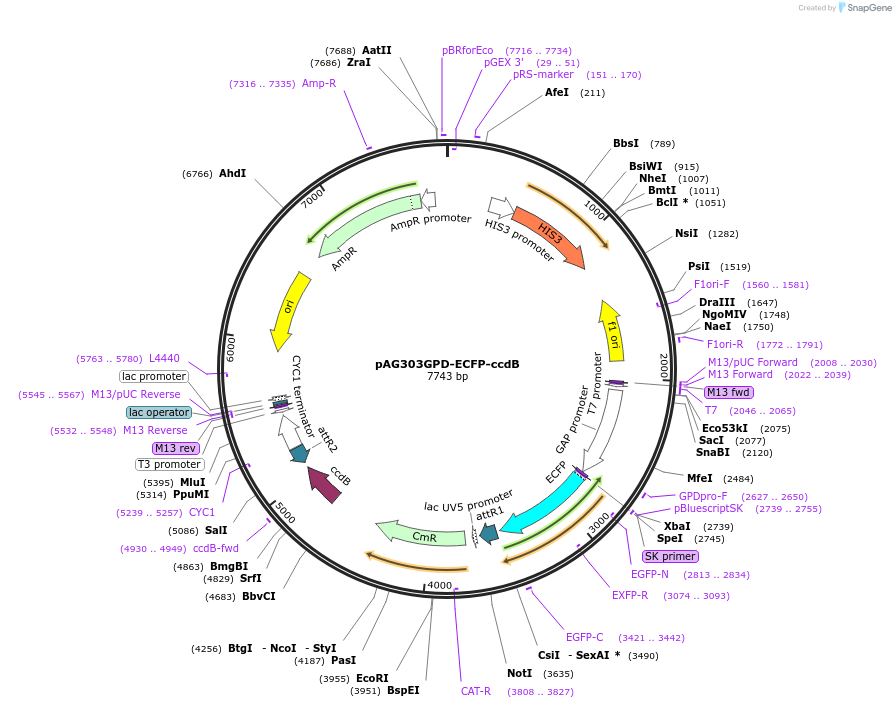
pAG303GPD-ECFP-ccdB
Plasmid#14278DepositorTypeEmpty backboneUseGateway destinationTagsECFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
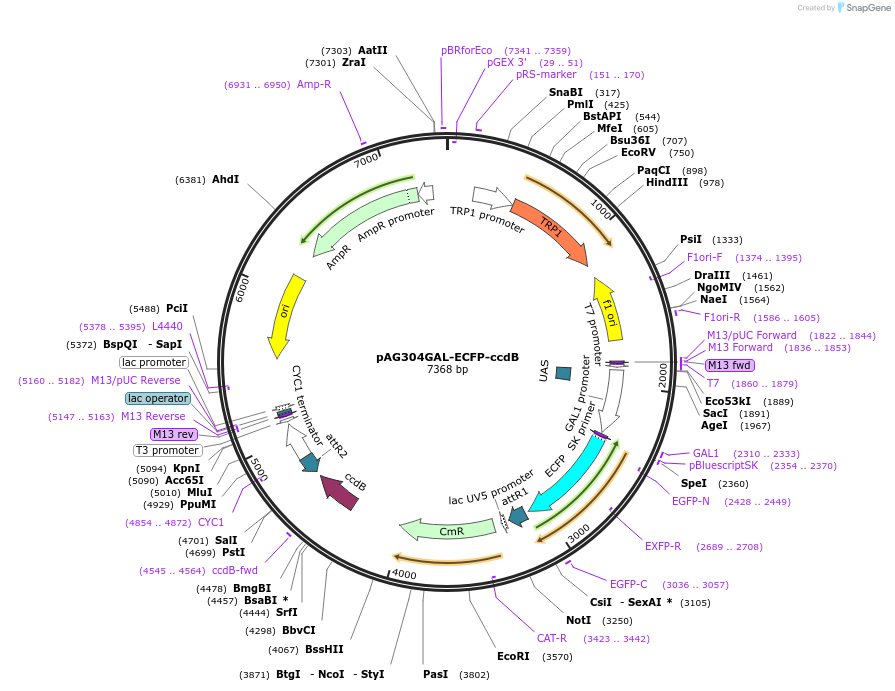
pAG304GAL-ECFP-ccdB
Plasmid#14279DepositorTypeEmpty backboneUseGateway destinationTagsECFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
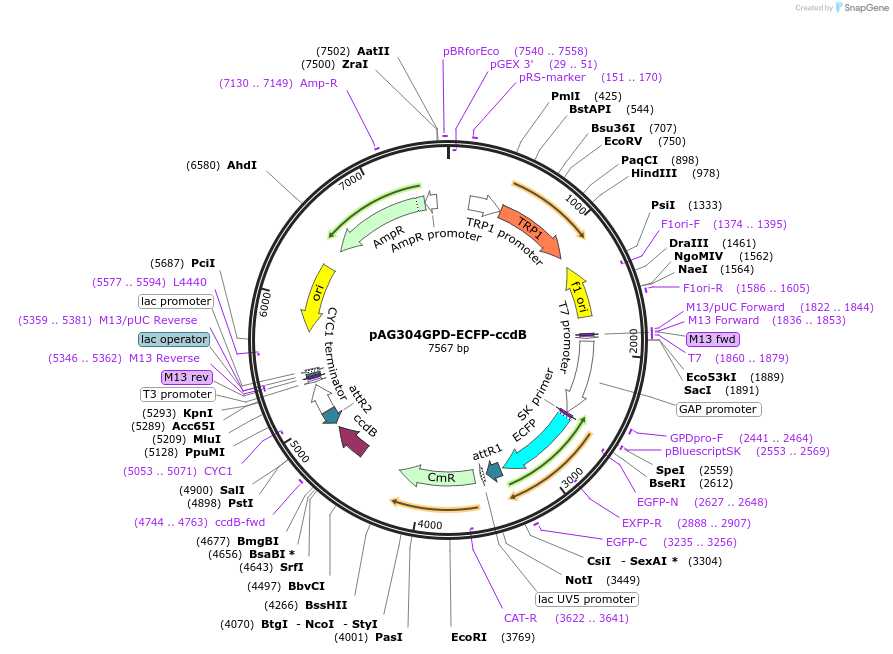
pAG304GPD-ECFP-ccdB
Plasmid#14280DepositorTypeEmpty backboneUseGateway destinationTagsECFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
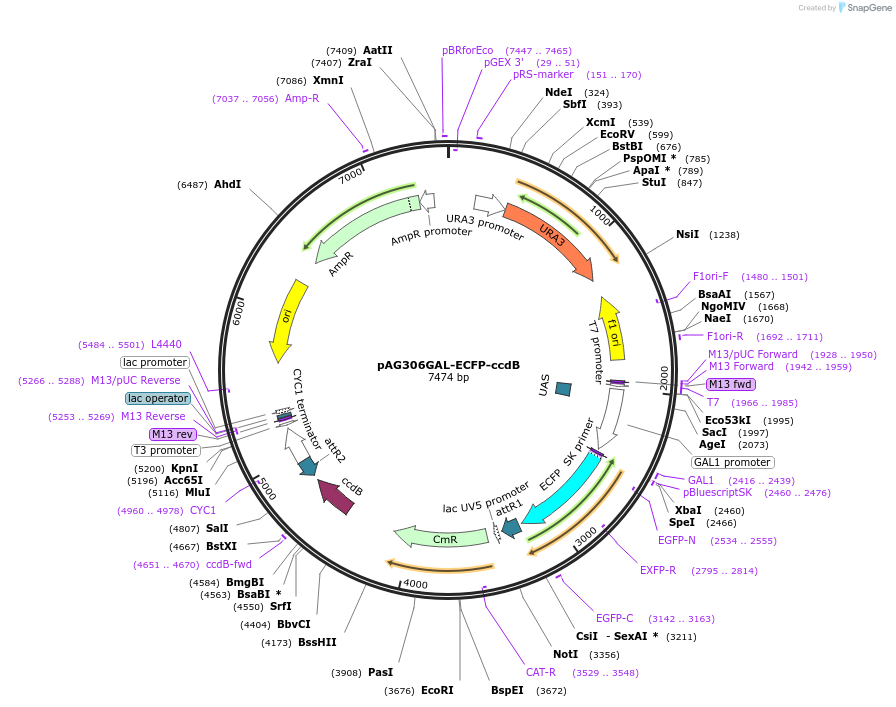
pAG306GAL-ECFP-ccdB
Plasmid#14283DepositorTypeEmpty backboneUseGateway destinationTagsECFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only -
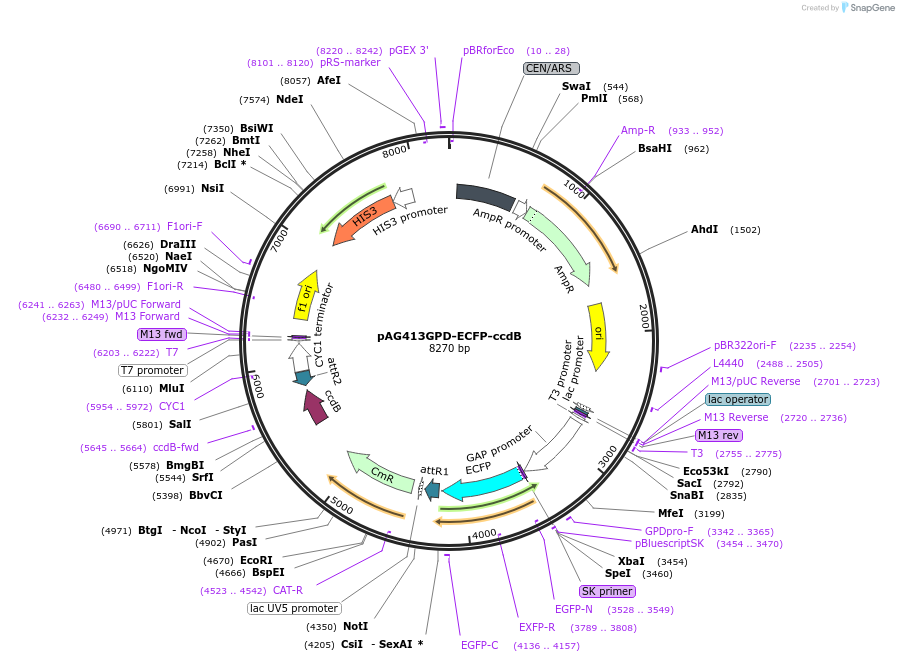
pAG413GPD-ECFP-ccdB
Plasmid#14286DepositorTypeEmpty backboneUseGateway destinationTagsECFPExpressionYeastAvailable SinceJune 18, 2007AvailabilityAcademic Institutions and Nonprofits only