We narrowed to 13,131 results for: LIC
-
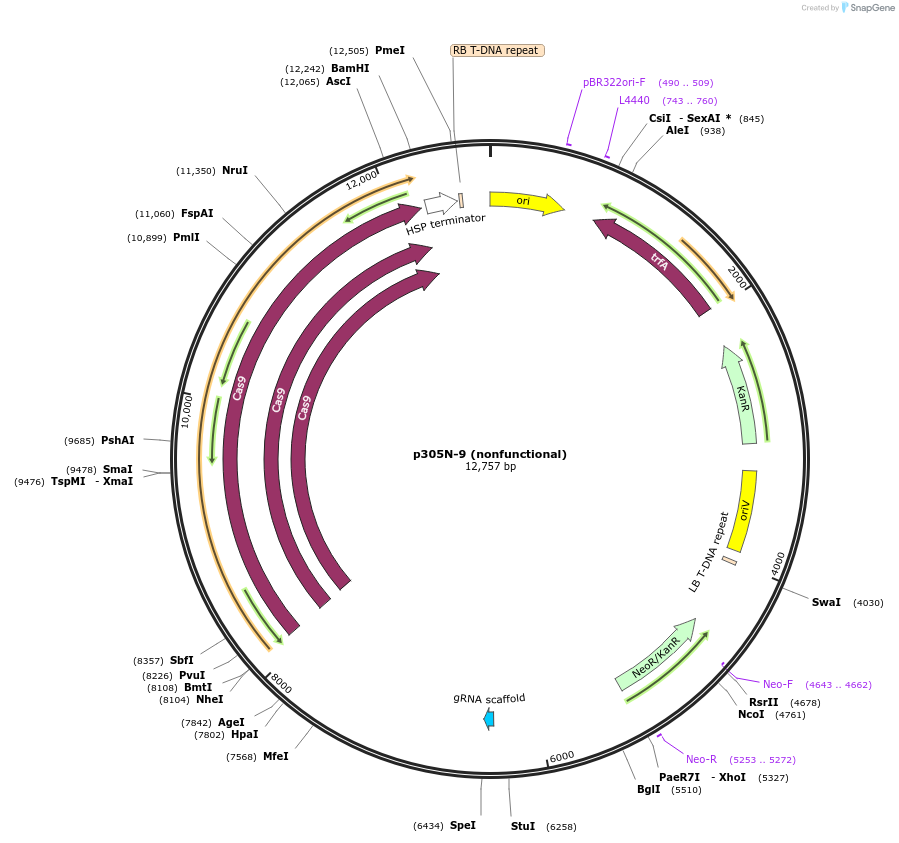
Plasmid#246314PurposeEvaluation of PtU6.2c4 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c4 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationdeletions: -A (-41 from TSS), -T (-30), -T (-29);…PromoterPtU6.2c4 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
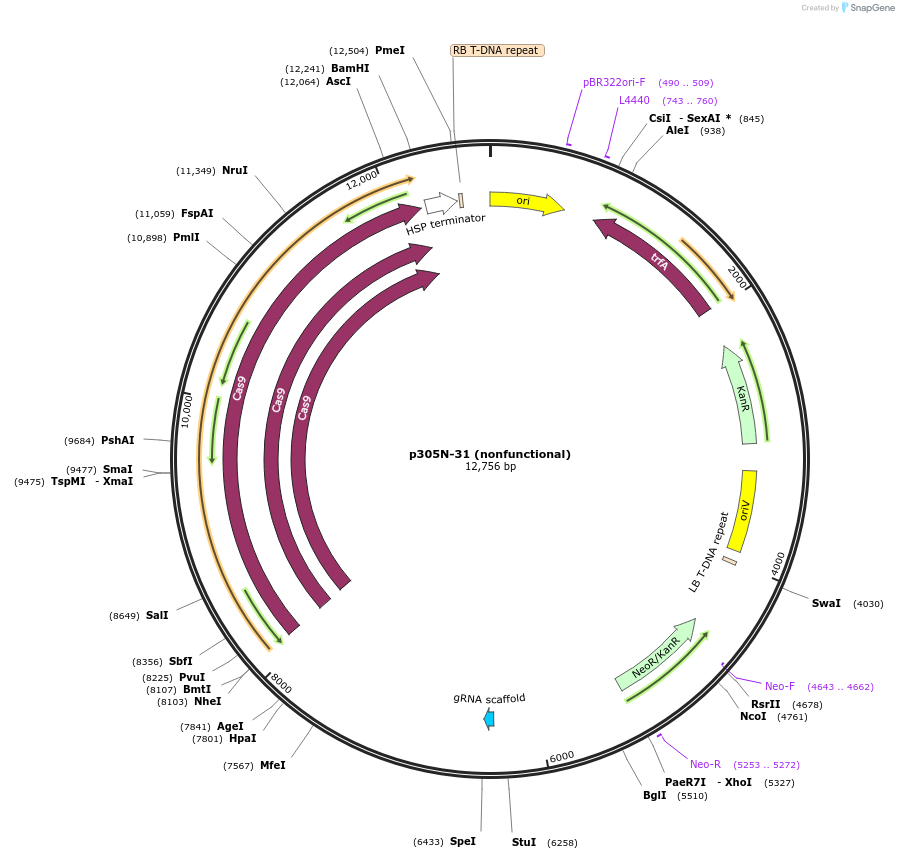
p305N-9 (nonfunctional)
Plasmid#246292PurposeEvaluation of AtU6.29c13 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.29c13 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationdeletions: -A (-58 from TSS), -T (-57) within USEPromoterAtU6.29c13 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
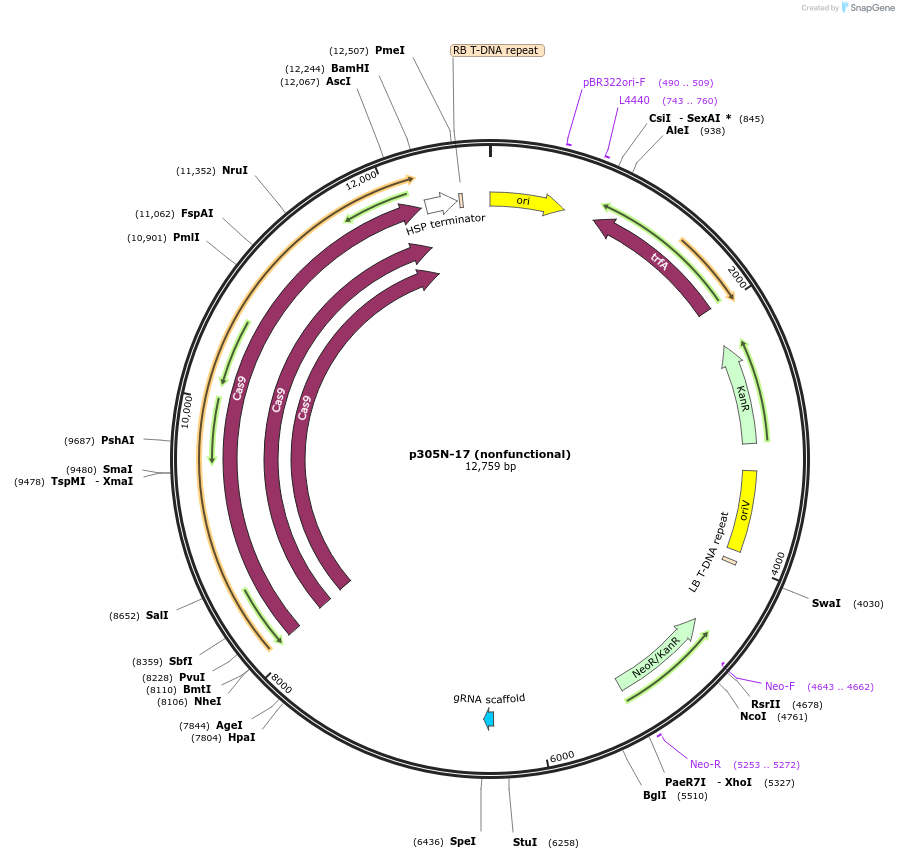
p305N-17 (nonfunctional)
Plasmid#246300PurposeEvaluation of HbU6.2m3 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertHbU6.2m3 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-T (-29 from TSS), G-to-A (-24)PromoterHbU6.2m3 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
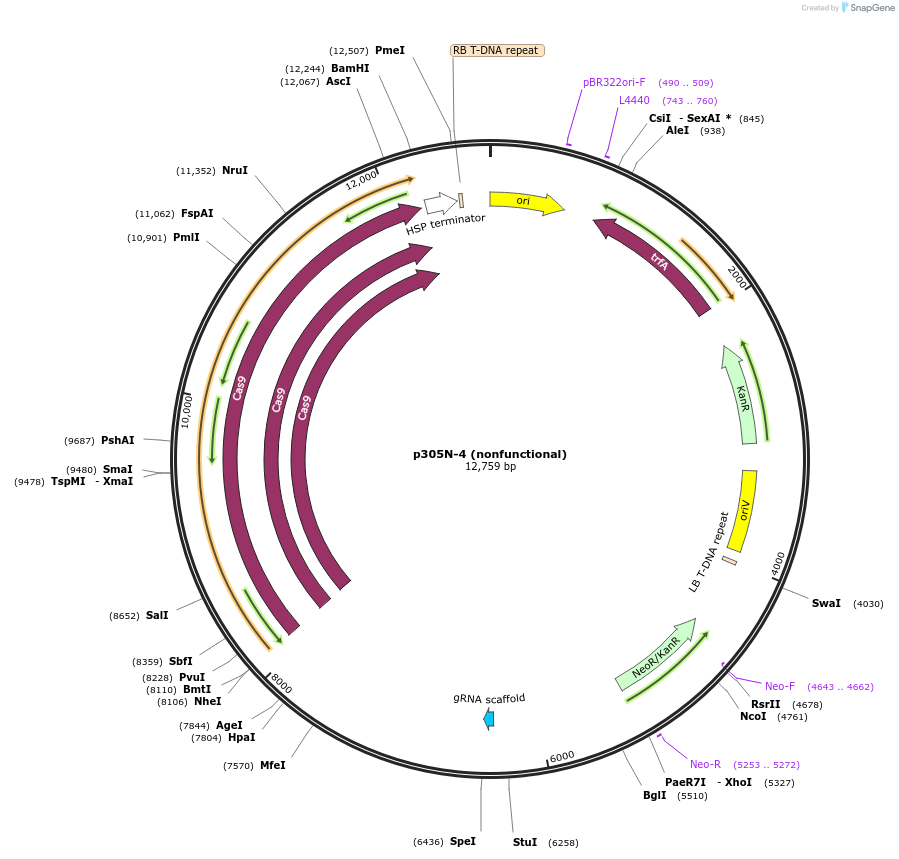
p305N-4 (nonfunctional)
Plasmid#246287PurposeEvaluation of AtU6.1c1 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.1c1 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationG-to-C at -15 from TSSPromoterAtU6.1c1 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
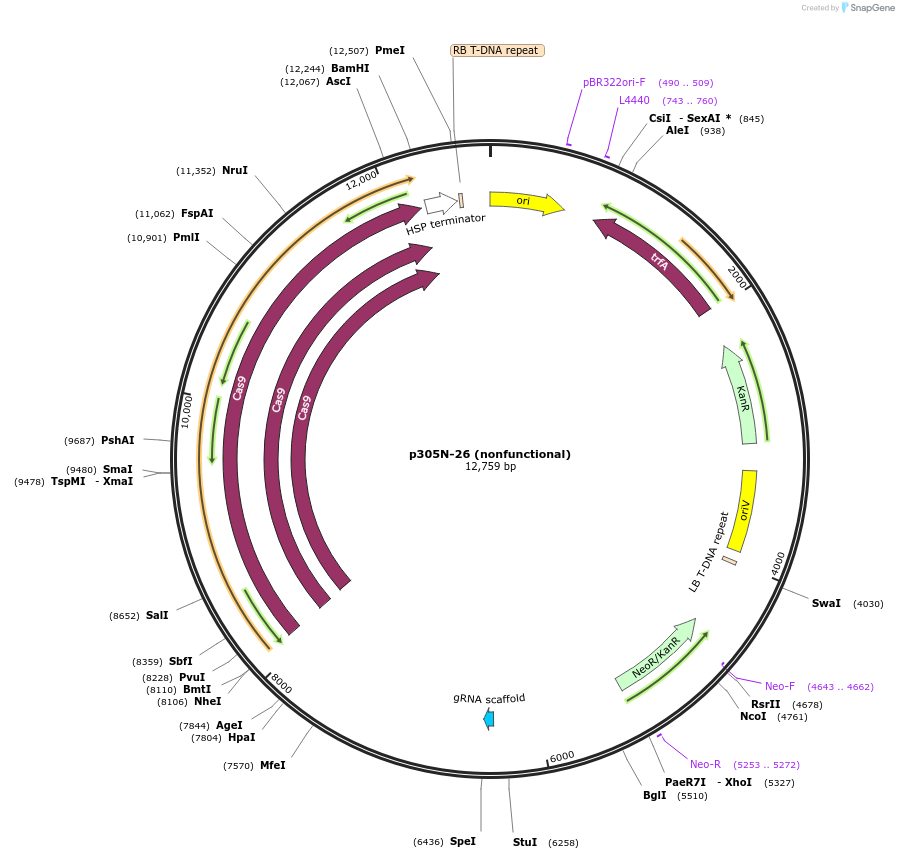
p305N-26 (nonfunctional)
Plasmid#246309PurposeEvaluation of MtU6.6m7 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertMtU6.6m7 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-A (-63 from TSS), T-to-G (-57)PromoterMtU6.6m7 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
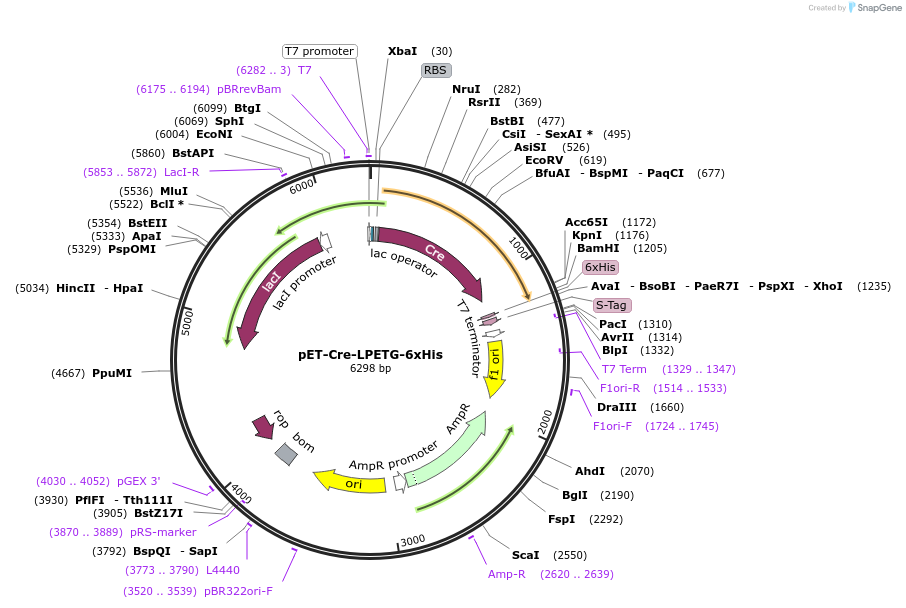
pET-Cre-LPETG-6xHis
Plasmid#237224PurposeExpresses LPETG-tagged Cre recombinase in bacteriaDepositorAvailable SinceJuly 29, 2025AvailabilityAcademic Institutions and Nonprofits only -
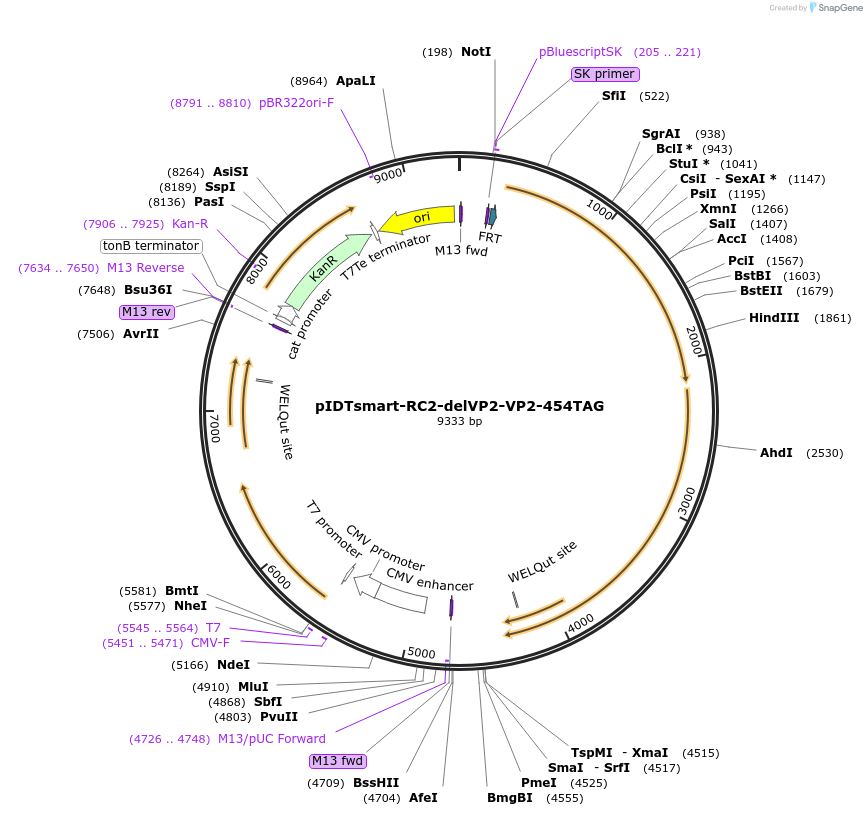
pIDTsmart-RC2-delVP2-VP2-454TAG
Plasmid#240060PurposeExpress AAV2 capsid with ncAA mutant at site 454 of VP2DepositorInsertsRC2-DelVP2
VP2-454-TAG
UseAAVExpressionMammalianAvailable SinceJune 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
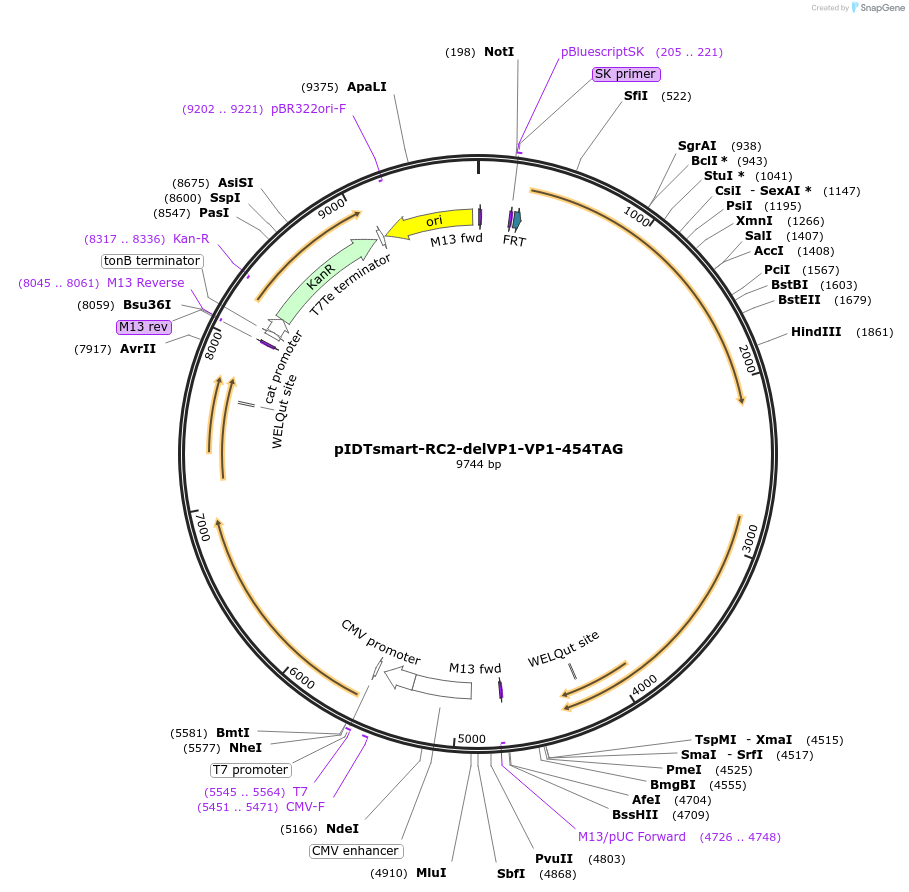
pIDTsmart-RC2-delVP1-VP1-454TAG
Plasmid#240059PurposeExpress AAV2 capsid with ncAA mutant at site 454 of VP1DepositorInsertsRC2-DelVP1
VP1-454-TAG
UseAAVExpressionMammalianAvailable SinceJune 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
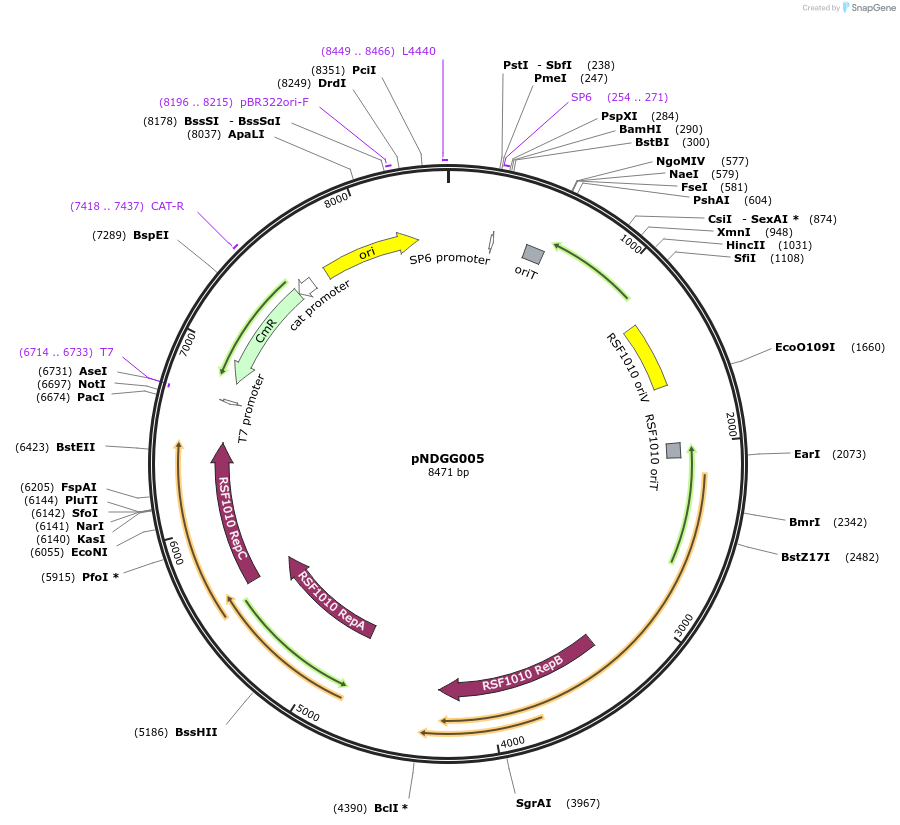
pNDGG005
Plasmid#231309PurposeVector part for BEVA; BEVA2.0 Position 3 RSF1010 broad host range origin of replication with oriT, CmR.DepositorInsertBEVA2.0 Position 3 RSF1010 broad host range origin of replication with oriT, CmR.
ExpressionBacterialAvailable SinceMay 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
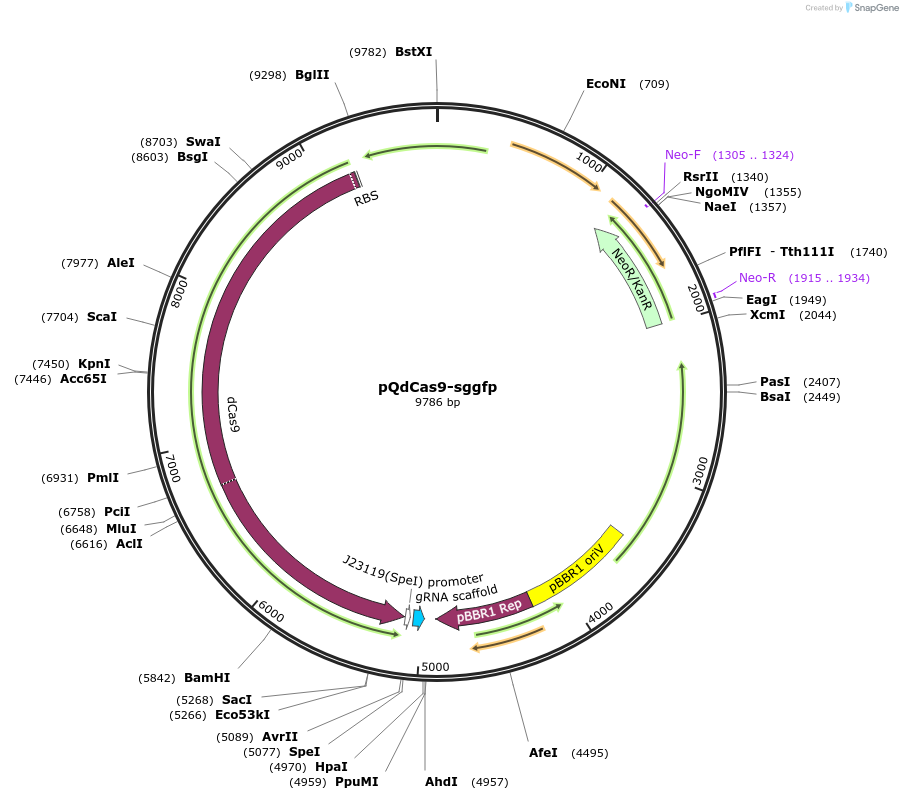
pQdCas9-sggfp
Plasmid#236184PurposeThe plasmid pQdCas9-sggfp expresses the dCas9 endonuclease and the sgRNA targeting the gfp gene.DepositorInsertQuorum sensing cassette luxI, luxR, and the LuxR-dependent promoter pLuxI , dCas9, and sgRNA for GFP gene
PromoterQuorum sensing promoterAvailable SinceApril 25, 2025AvailabilityAcademic Institutions and Nonprofits only