We narrowed to 167,150 results for: htt
-
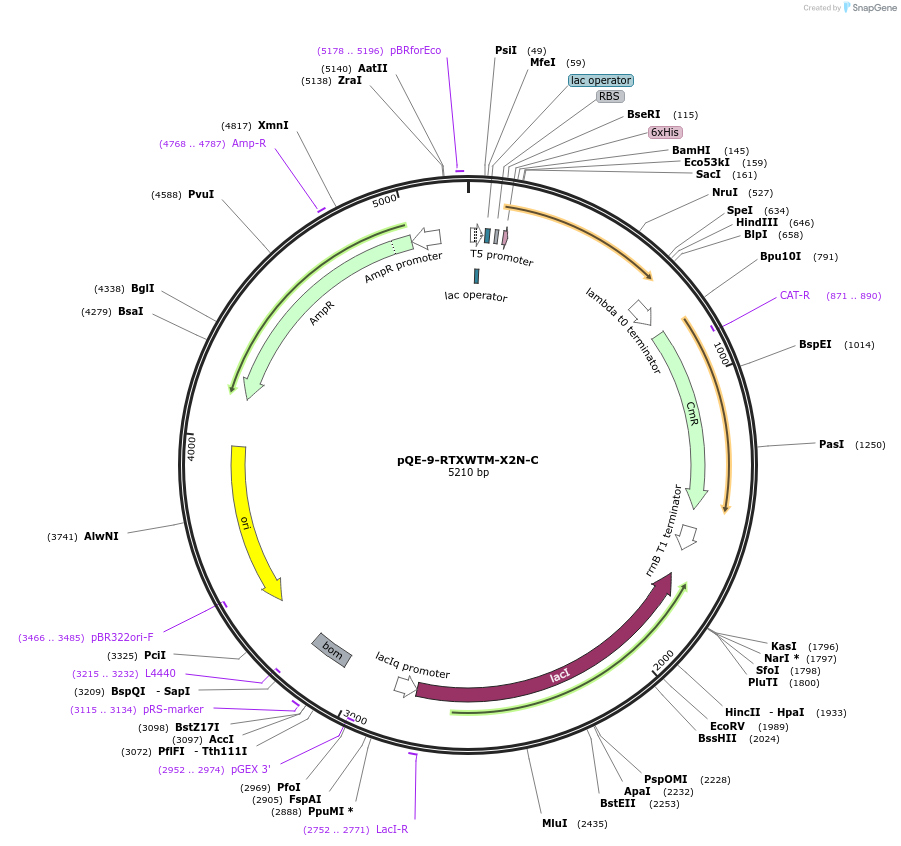
Plasmid#225968PurposeBlock V Repeats-In-Toxin (RTX) protein with global substitution of asparagine in nonapeptide repeats and C-terminal Capping DomainDepositorInsertRTXWTM-X2N-C
Tags6xHisExpressionBacterialAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
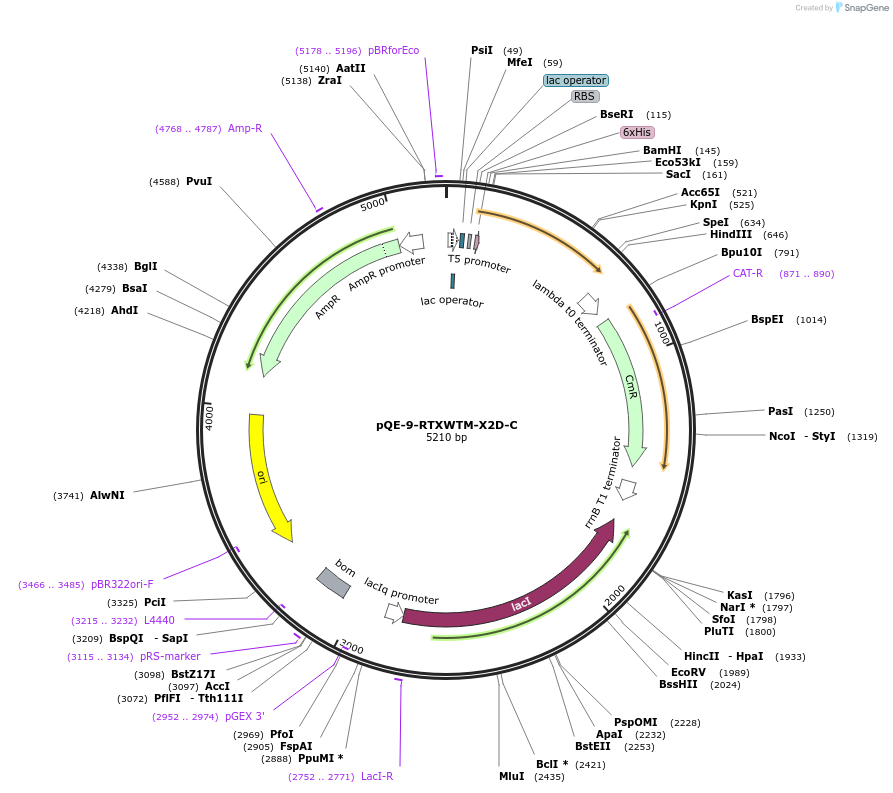
pQE-9-RTXWTM-X2D-C
Plasmid#225969PurposeBlock V Repeats-In-Toxin (RTX) protein with global substitution of aspartic acid in nonapeptide repeats and C-terminal Capping DomainDepositorInsertRTXWTM-X2D-C
Tags6xHisExpressionBacterialAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
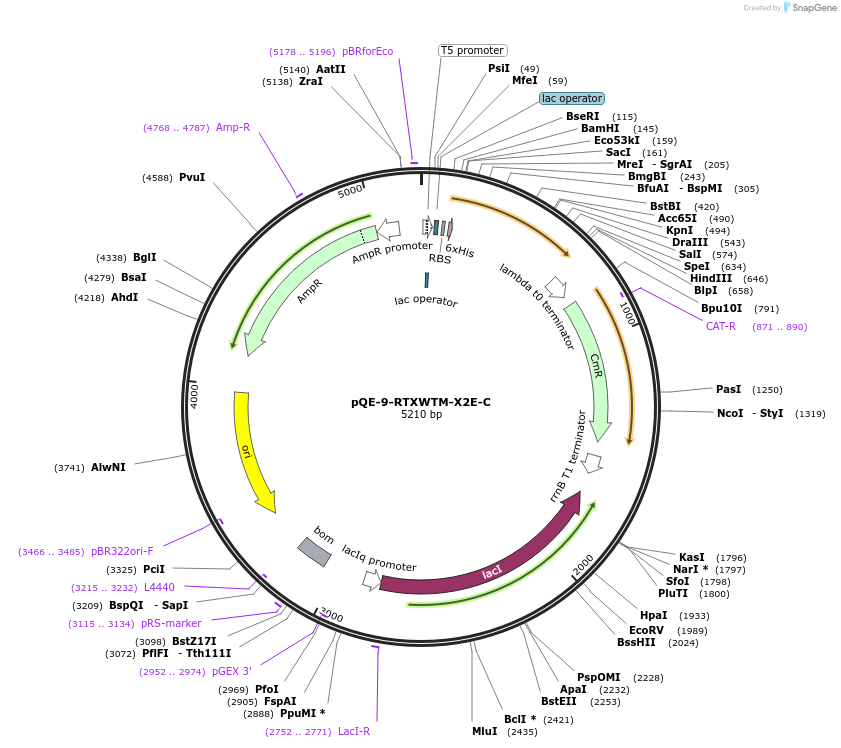
pQE-9-RTXWTM-X2E-C
Plasmid#225970PurposeBlock V Repeats-In-Toxin (RTX) protein with global substitution of glutamic acid in nonapeptide repeats and C-terminal Capping DomainDepositorInsertRTXWTM-X2E-C
Tags6xHisExpressionBacterialAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
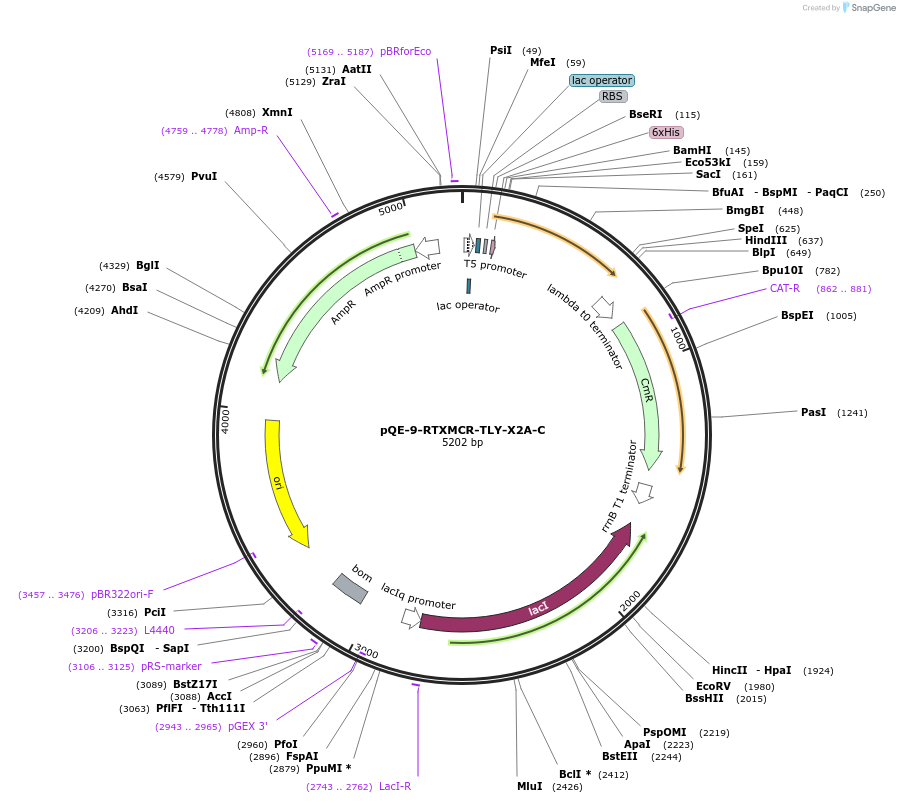
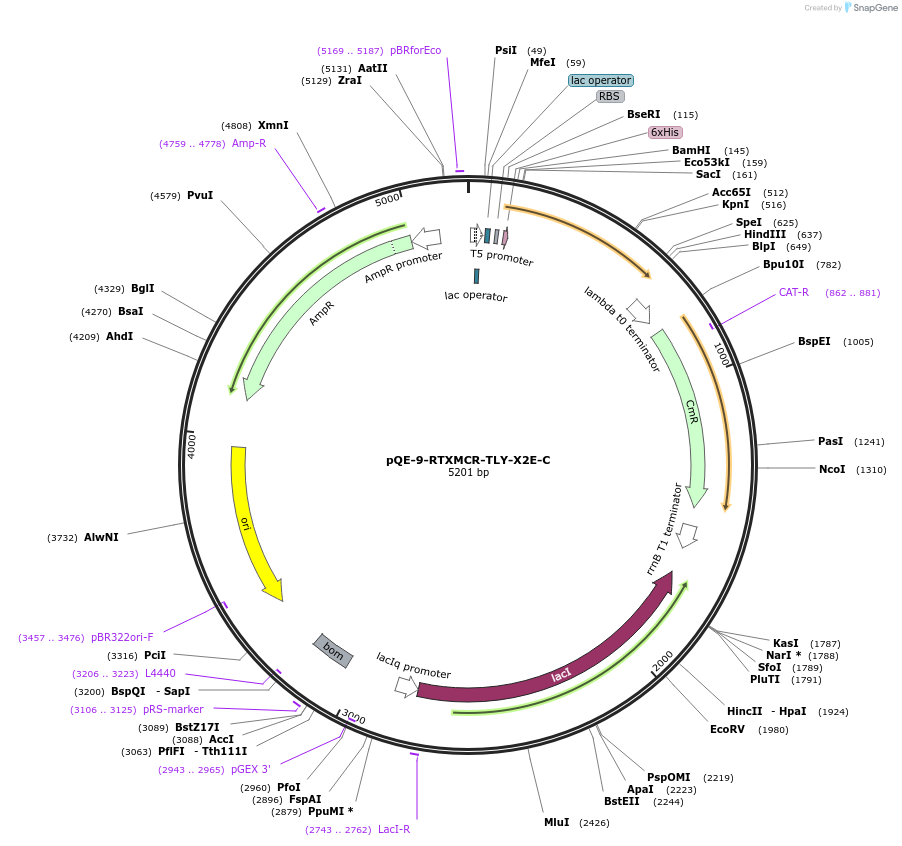
pQE-9-RTXMCR-TLY-X2A-C
Plasmid#225971PurposeRepeats-In-Toxin (RTX) protein consensus sequence GGAGADTLY repeated nine times with C-terminal Capping DomainDepositorInsertRTMCR-TLY-X2A-C
Tags6xHisExpressionBacterialAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
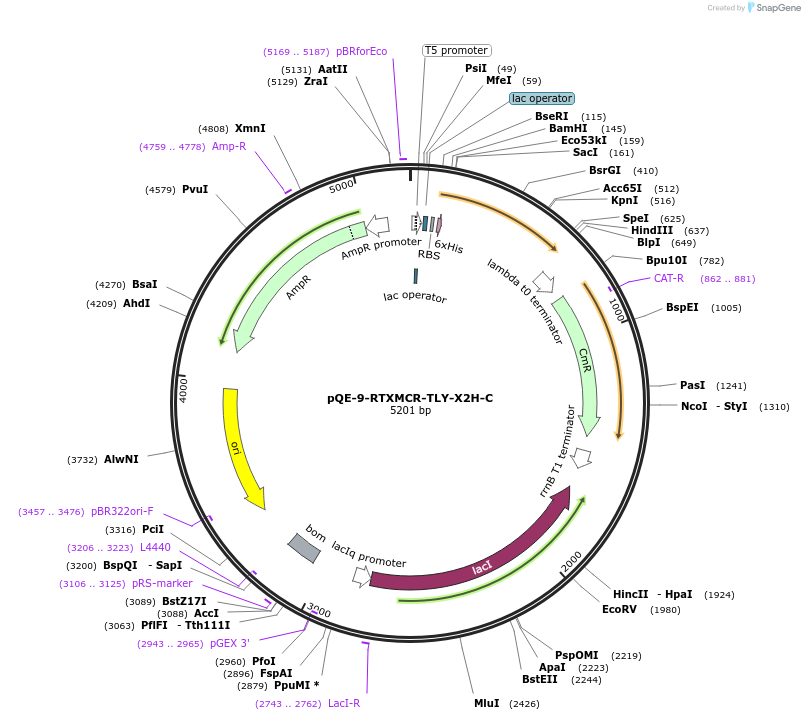
pQE-9-RTXMCR-TLY-X2H-C
Plasmid#225972PurposeRepeats-In-Toxin (RTX) protein consensus sequence GGAGHDTLY repeated nine times with C-terminal Capping DomainDepositorInsertRTXMCR-TLY-X2H-C
Tags6xHisExpressionBacterialAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
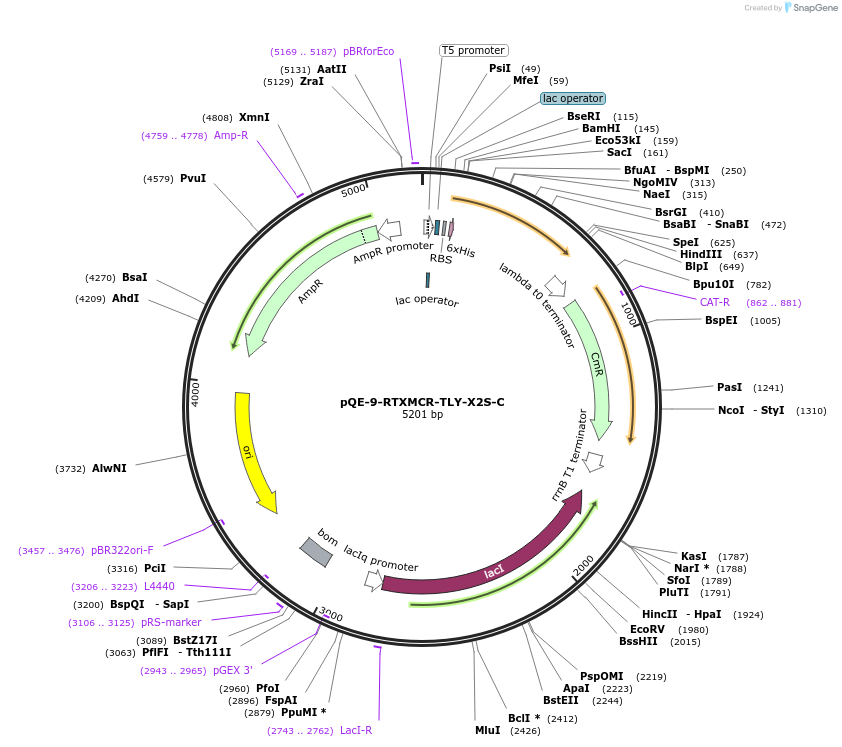
pQE-9-RTXMCR-TLY-X2S-C
Plasmid#225973PurposeRepeats-In-Toxin (RTX) protein consensus sequence GGAGSDTLY repeated nine times with C-terminal Capping DomainDepositorInsertRTXMCR-TLY-X2S-C
Tags6xHisExpressionBacterialAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
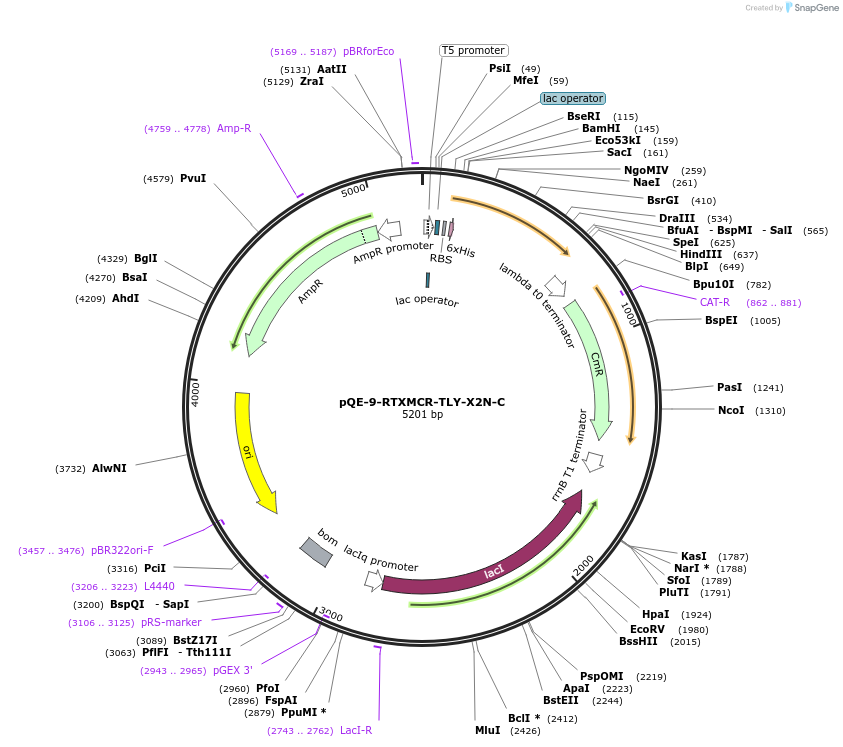
pQE-9-RTXMCR-TLY-X2N-C
Plasmid#225974PurposeRepeats-In-Toxin (RTX) protein consensus sequence GGAGNDTLY repeated nine times with C-terminal Capping DomainDepositorInsertRTXMCR-TLY-X2N-C
Tags6xHisExpressionBacterialAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
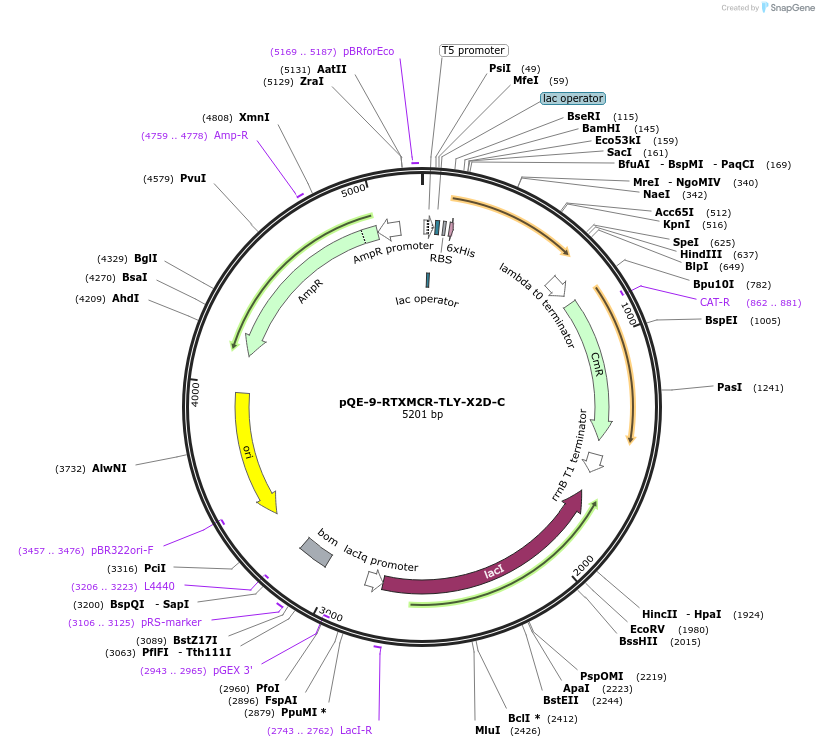
pQE-9-RTXMCR-TLY-X2D-C
Plasmid#225975PurposeRepeats-In-Toxin (RTX) protein consensus sequence GGAGDDTLY repeated nine times with C-terminal Capping DomainDepositorInsertRTXMCR-TLY-X2D-C
Tags6xHisExpressionBacterialAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
pQE-9-RTXMCR-TLY-X2E-C
Plasmid#225976PurposeRepeats-In-Toxin (RTX) protein consensus sequence GGAGEDTLY repeated nine times with C-terminal Capping DomainDepositorInsertRTXMCR-TLY-X2E-C
Tags6xHisExpressionBacterialAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
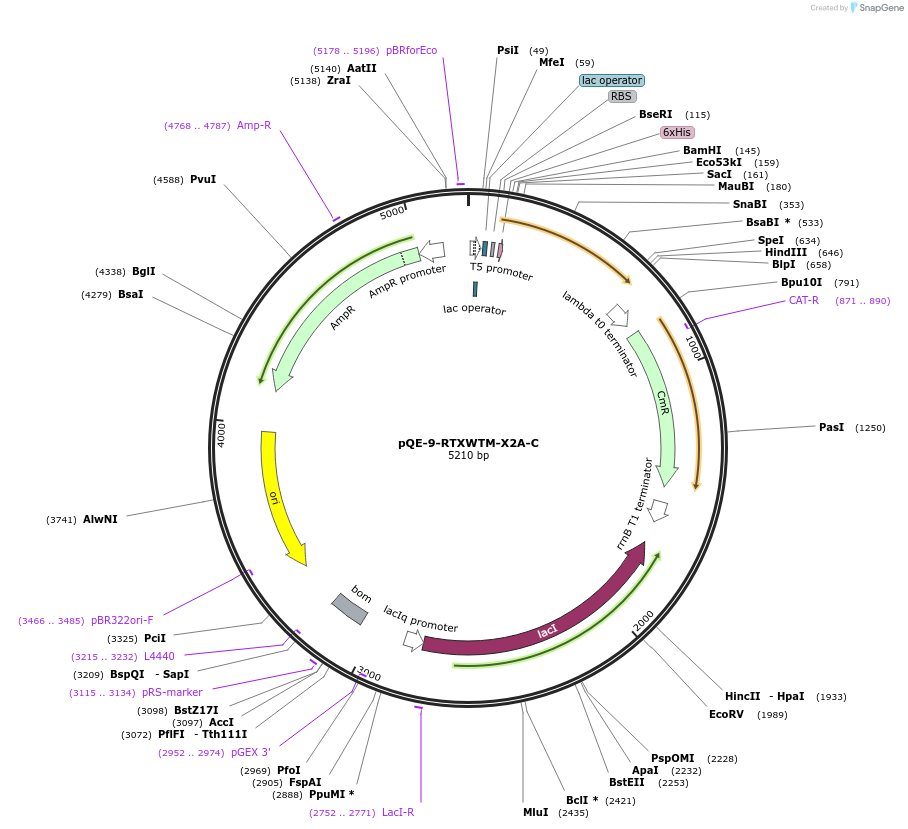
pQE-9-RTXWTM-X2A-C
Plasmid#225965PurposeBlock V Repeats-In-Toxin (RTX) protein with global substitution of alanine in nonapeptide repeats and C-terminal Capping DomainDepositorInsertRTXWTM-X2A-C
Tags6xHisExpressionBacterialAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
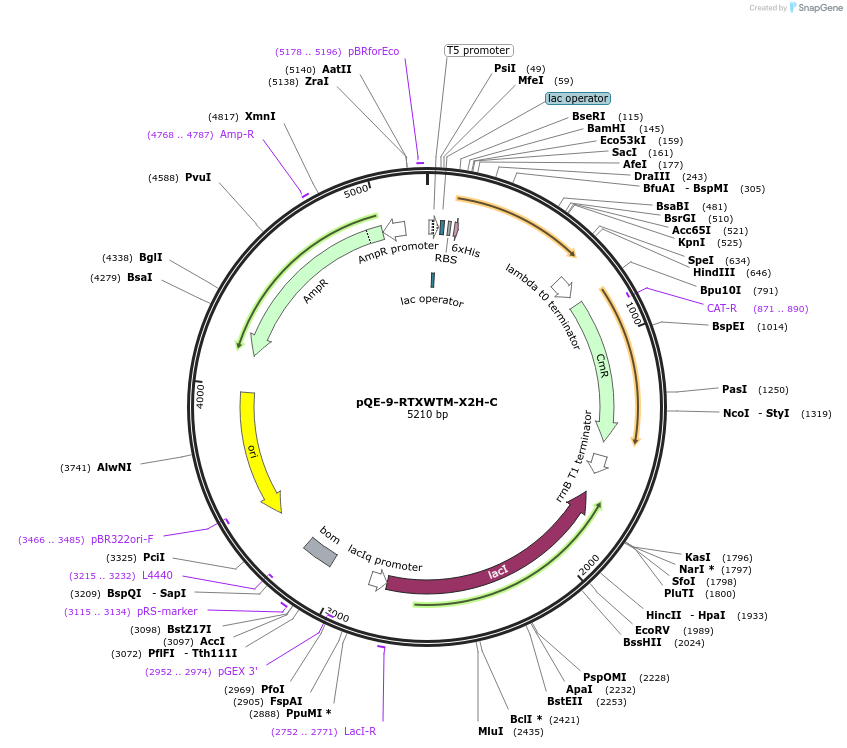
pQE-9-RTXWTM-X2H-C
Plasmid#225966PurposeBlock V Repeats-In-Toxin (RTX) protein with global substitution of histidine in nonapeptide repeats and C-terminal Capping DomainDepositorInsertRTXWTM-X2H-C
Tags6xHisExpressionBacterialAvailable SinceNov. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
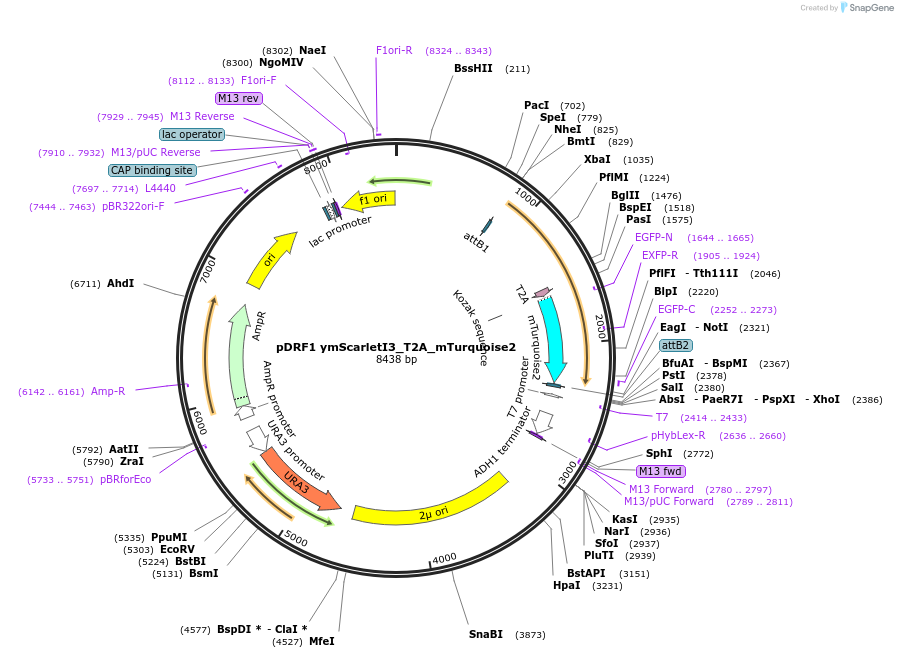
pDRF1 ymScarletI3_T2A_mTurquoise2
Plasmid#219829Purposeequimolar expression of ymScarletI3 and mTurquoise2 in budding yeastDepositorInsertymScarletI3_T2A_mTurquoise2
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
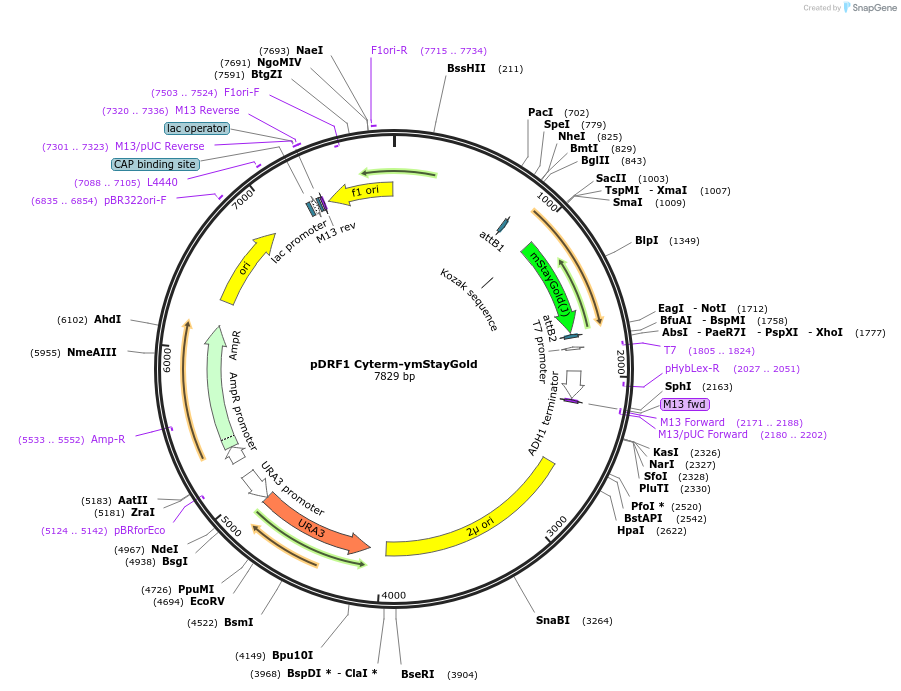
pDRF1 Cyterm-ymStayGold
Plasmid#219855Purposeexpression of Cyterm-ymStayGoldd in budding yeastDepositorInsertCyterm-ymStayGold
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
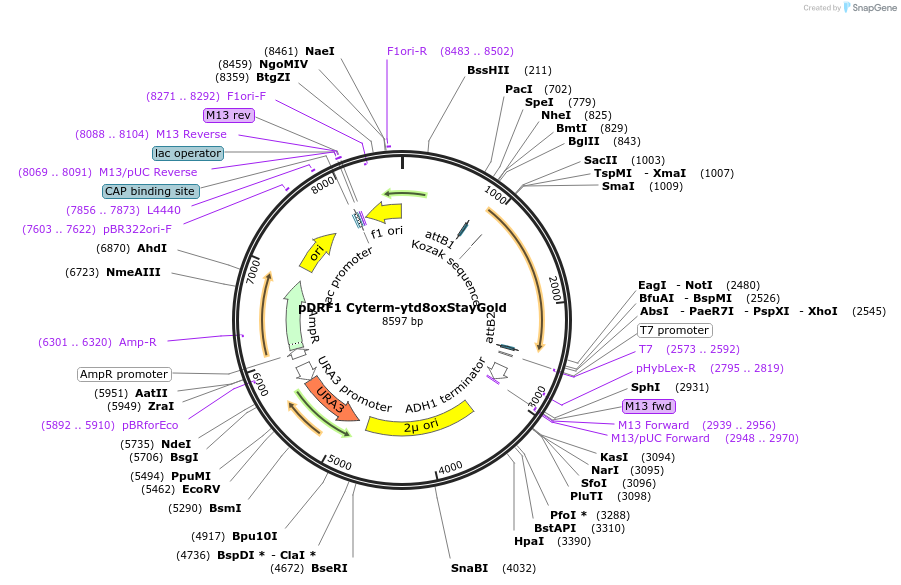
pDRF1 Cyterm-ytd8oxStayGold
Plasmid#219853Purposeexpression of Cyterm-ytd8oxStayGold in budding yeastDepositorInsertCyterm-ytd8oxStayGold
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
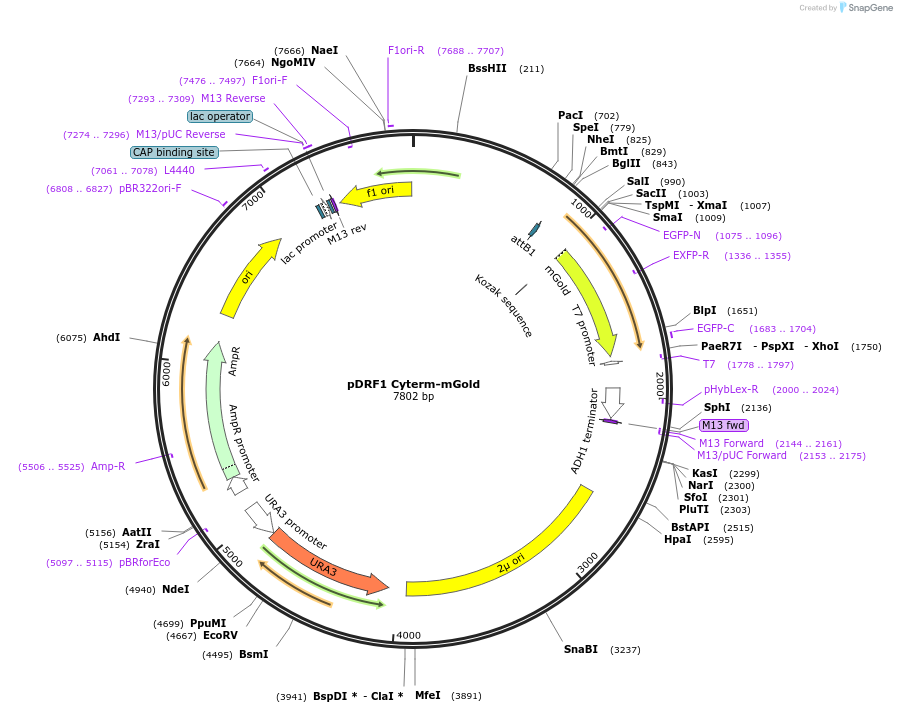
pDRF1 Cyterm-mGold
Plasmid#224084PurposeExpresses Cyterm-mGold to check for monomericity.DepositorInsertCyterm-mGold
ExpressionYeastAvailable SinceAug. 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
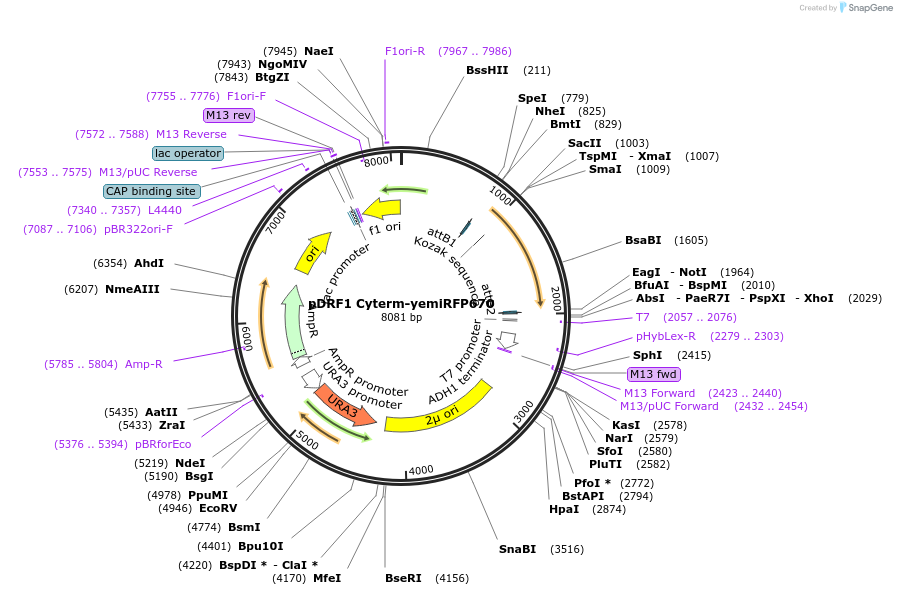
pDRF1 Cyterm-yemiRFP670
Plasmid#219856Purposeexpression of Cyterm-yemiRFP670 in budding yeastDepositorInsertCyterm-yemiRFP670
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
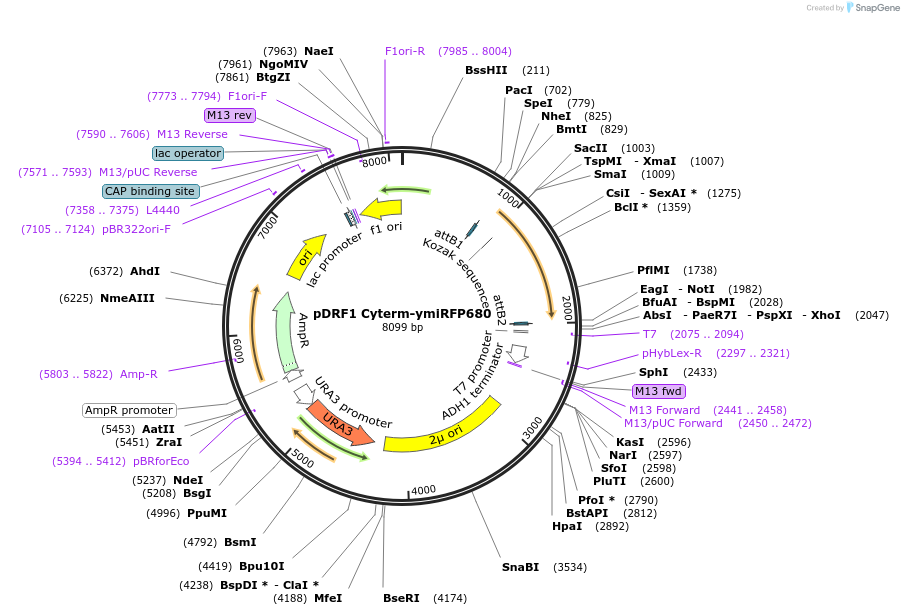
pDRF1 Cyterm-ymiRFP680
Plasmid#219854Purposeexpression of Cyterm-ymiRFP680 in budding yeastDepositorInsertCyterm-ymiRFP680
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
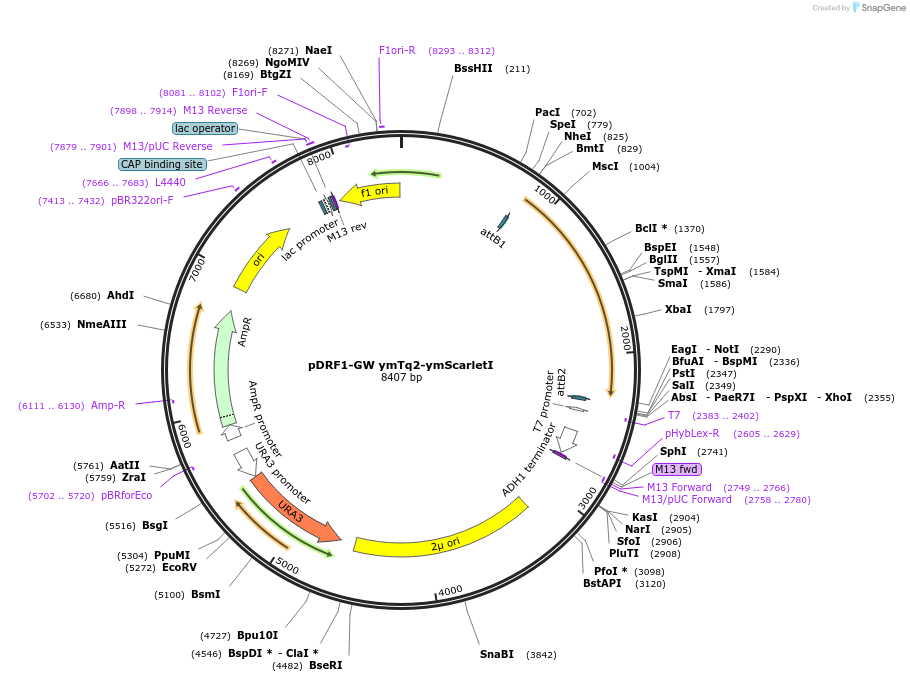
pDRF1-GW ymTq2-ymScarletI
Plasmid#205512PurposeExpression of ymTq2-ymScarletl in yeastDepositorInsertymTq2-ymScarletl
ExpressionYeastAvailable SinceSept. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
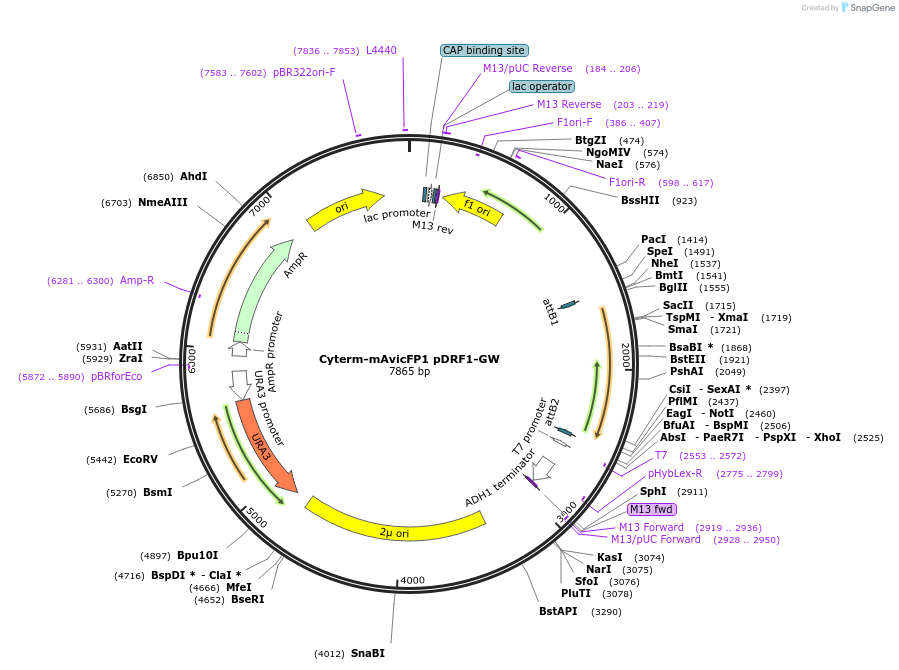
Cyterm-mAvicFP1 pDRF1-GW
Plasmid#168092PurposeExpresses Cyterm-mAvicFP1 to check for monomericity.DepositorInsertmAvicFP1
ExpressionYeastAvailable SinceJune 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
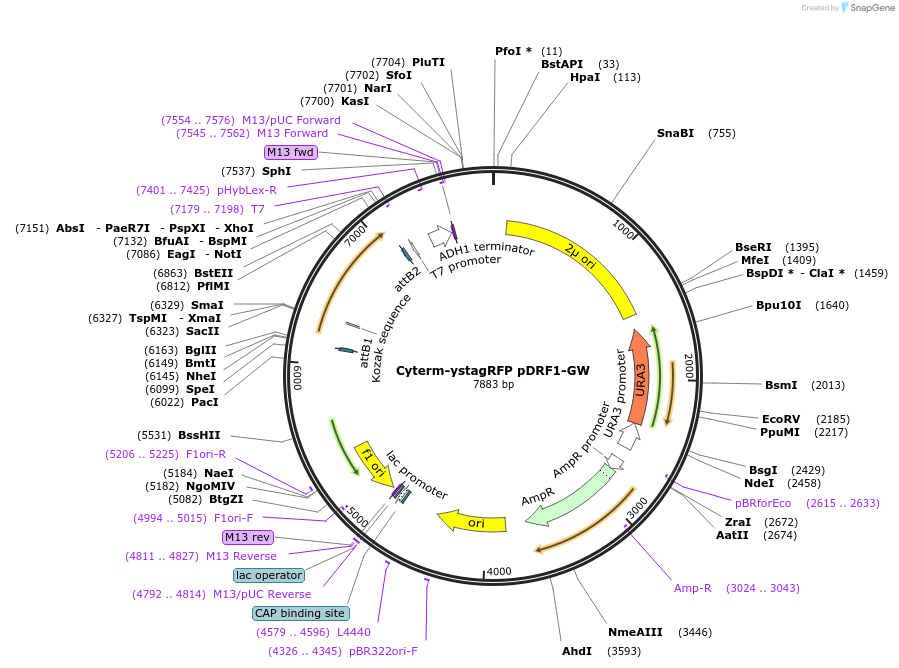
Cyterm-ystagRFP pDRF1-GW
Plasmid#168061PurposeExpresses Cyterm-ystagRFP to check for monomericity.DepositorInsertCyterm-ystagRFP
ExpressionYeastAvailable SinceJune 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
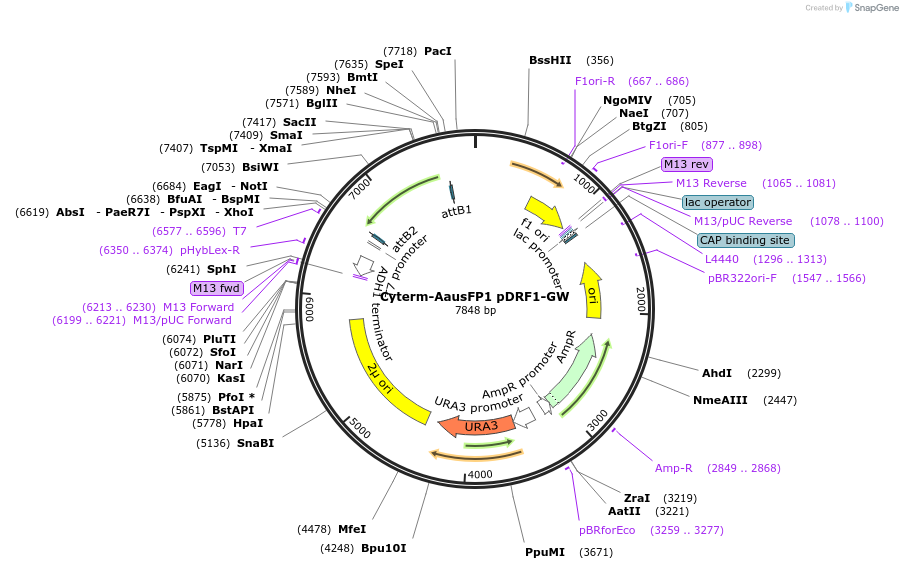
Cyterm-AausFP1 pDRF1-GW
Plasmid#168062PurposeExpresses Cyterm-AausFP1 to check for monomericity.DepositorInsertCyterm-AausFP1
ExpressionYeastAvailable SinceJune 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
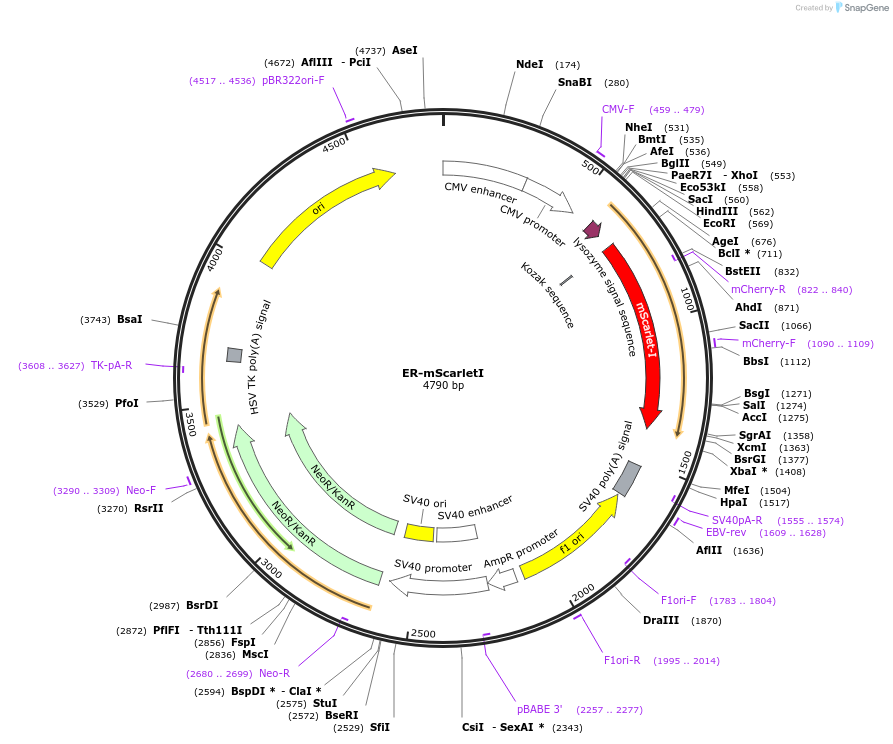
ER-mScarletI
Plasmid#137805PurposeVisualization of the Endoplasmic ReticulumDepositorInsertKDEL
ExpressionMammalianPromoterCMVAvailable SinceFeb. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
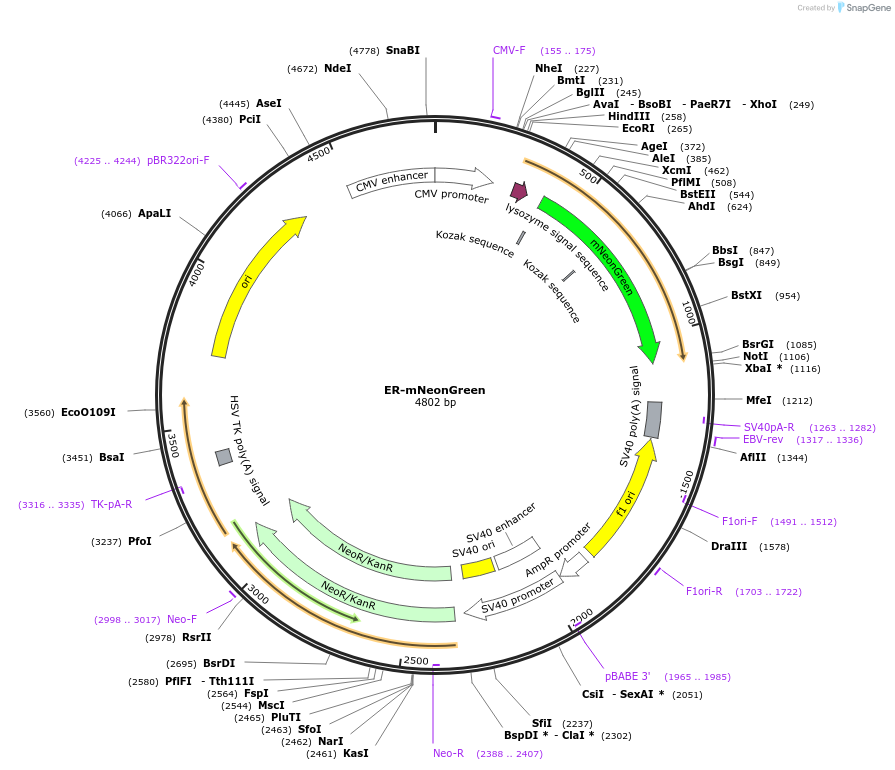
ER-mNeonGreen
Plasmid#137804PurposeVisualization of the Endoplasmic ReticulumDepositorInsertKDEL
ExpressionMammalianPromoterCMVAvailable SinceFeb. 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
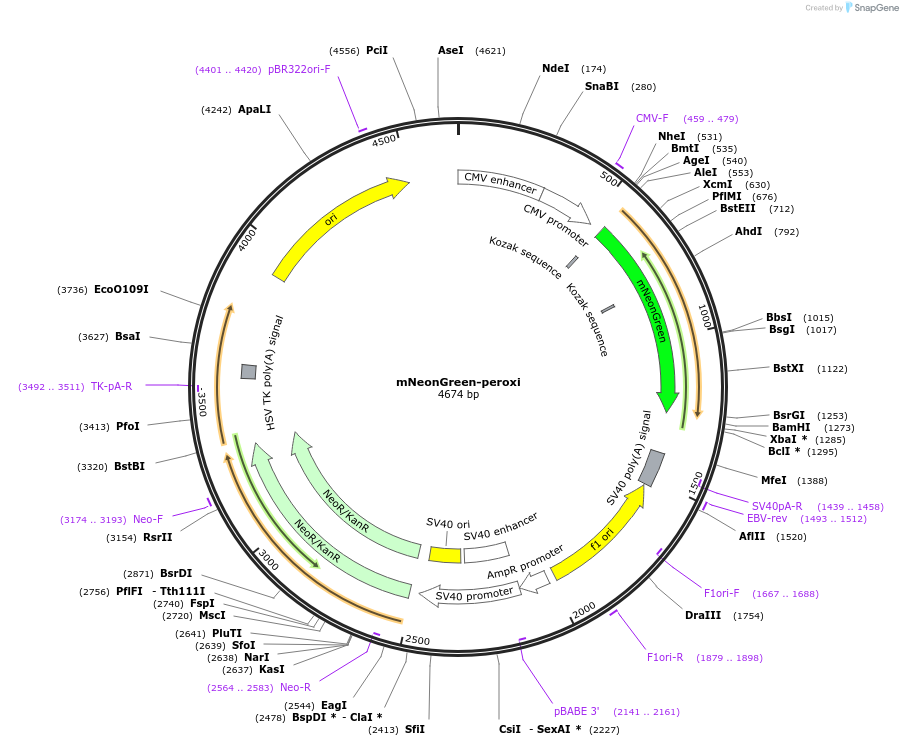
mNeonGreen-peroxi
Plasmid#137806PurposeVisualization of peroxisomesDepositorInsertSKL
ExpressionMammalianPromoterCMVAvailable SinceFeb. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
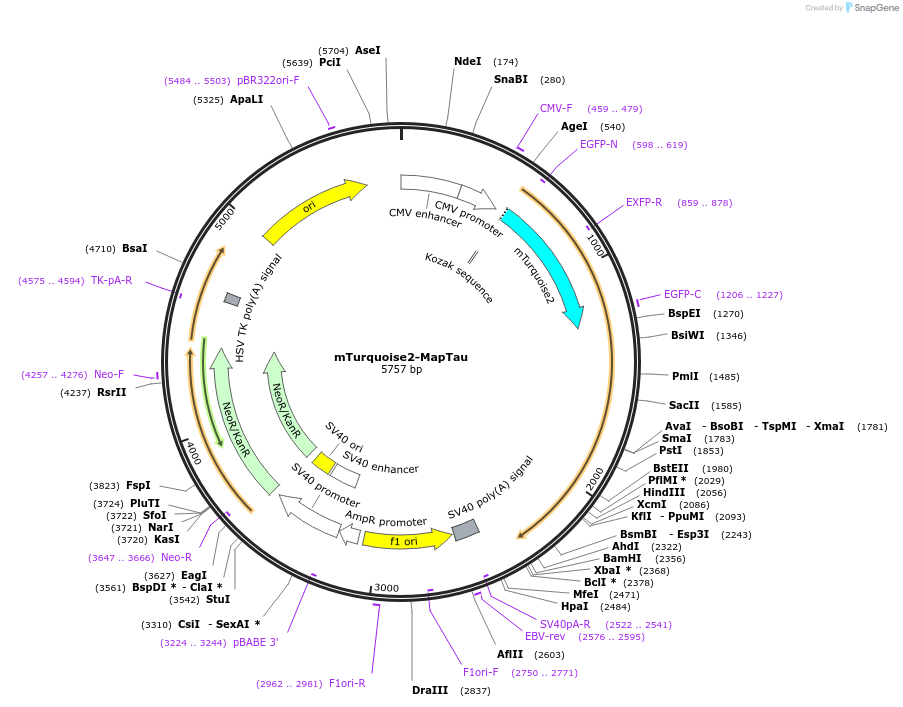
mTurquoise2-MapTau
Plasmid#137807PurposeVisualization of microtubulesDepositorInsertMapTau
ExpressionMammalianPromoterCMVAvailable SinceFeb. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
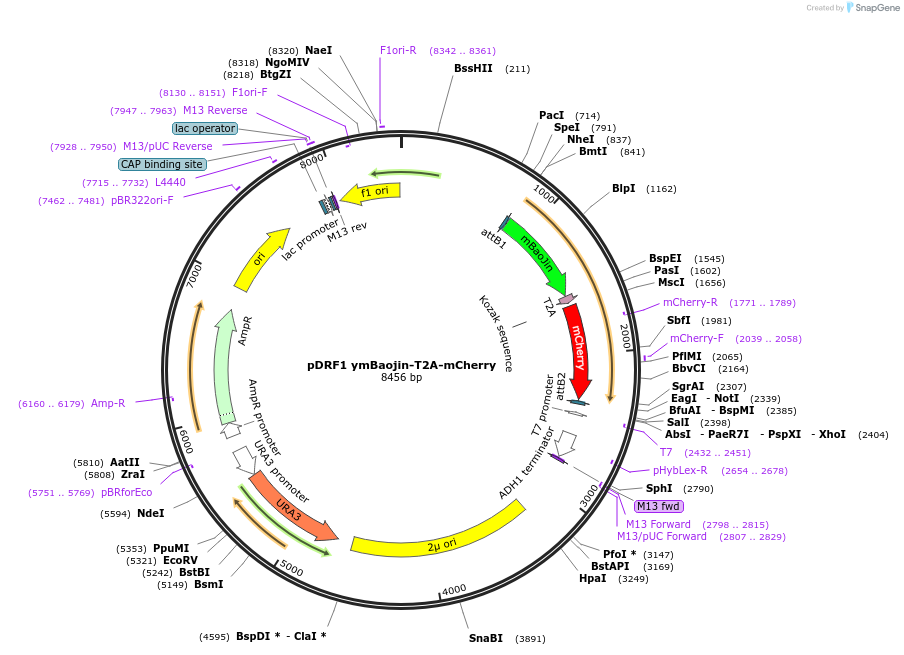
pDRF1 ymBaojin-T2A-mCherry
Plasmid#219857Purposeequimolar expression of ymBaojin and mCherry in budding yeastDepositorInsertymBaojin-T2A-mCherry
ExpressionYeastAvailable SinceAug. 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
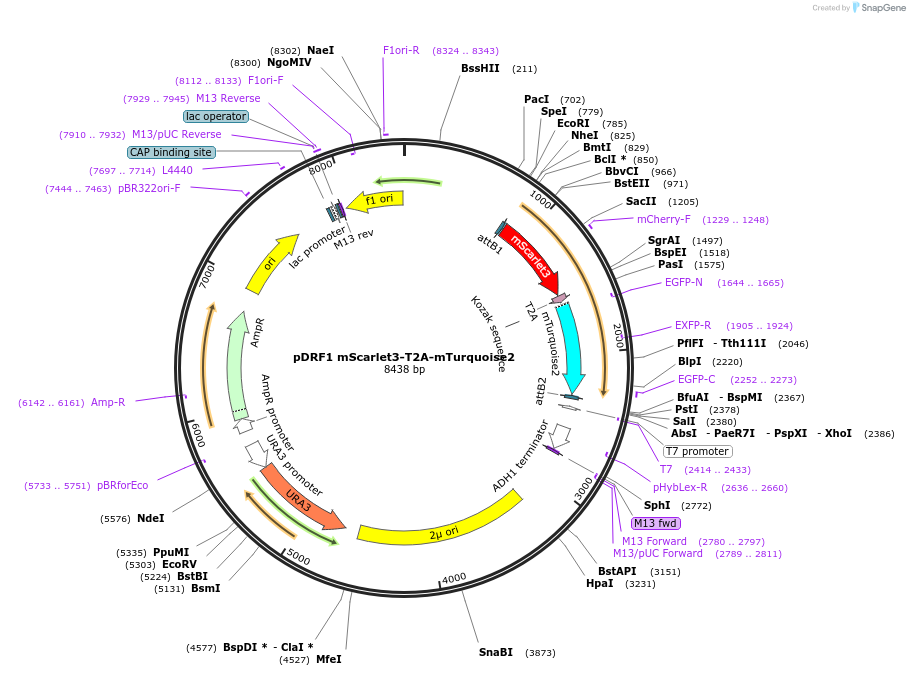
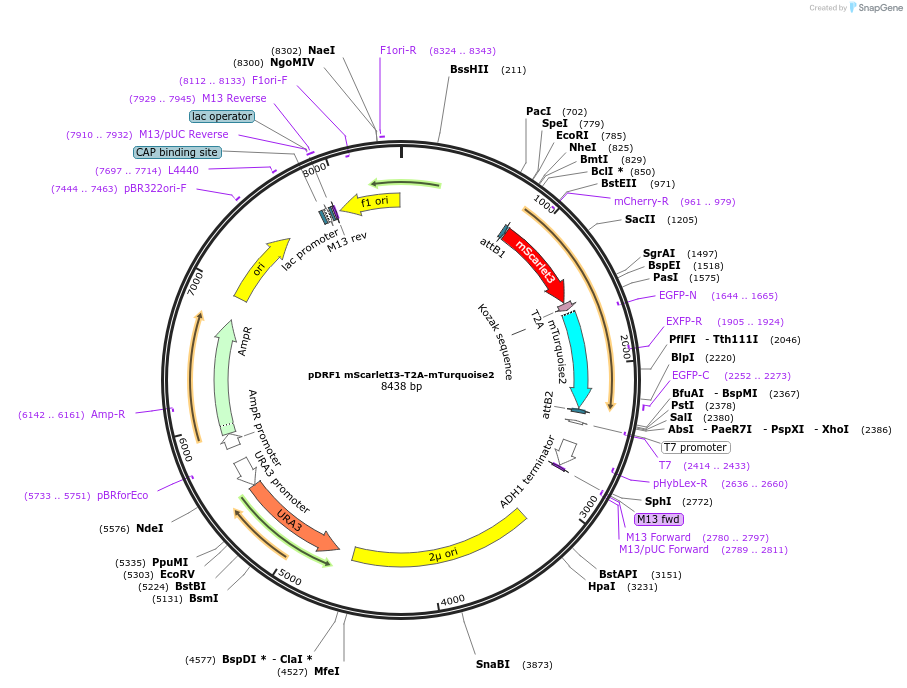
pDRF1 mScarlet3-T2A-mTurquoise2
Plasmid#219848Purposeequimolar expression of mScarlet3 and mTurquoise2 in budding yeastDepositorInsertmScarlet3-T2A-mTurquoise2
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
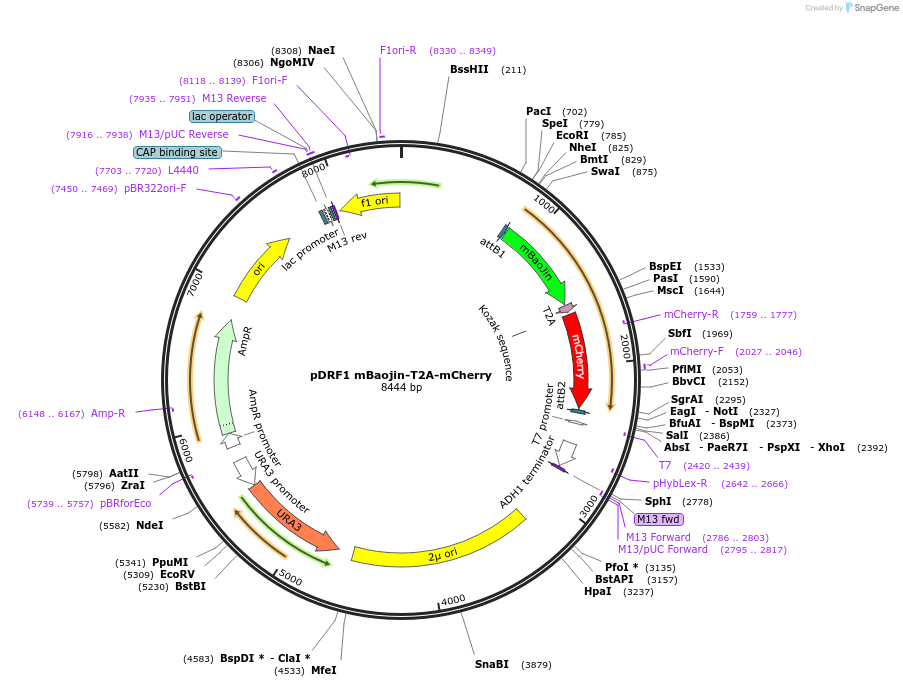
pDRF1 mBaojin-T2A-mCherry
Plasmid#219842Purposeequimolar expression of mBaojin and mCherry in budding yeastDepositorInsertmBaojin-T2A-mCherry
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
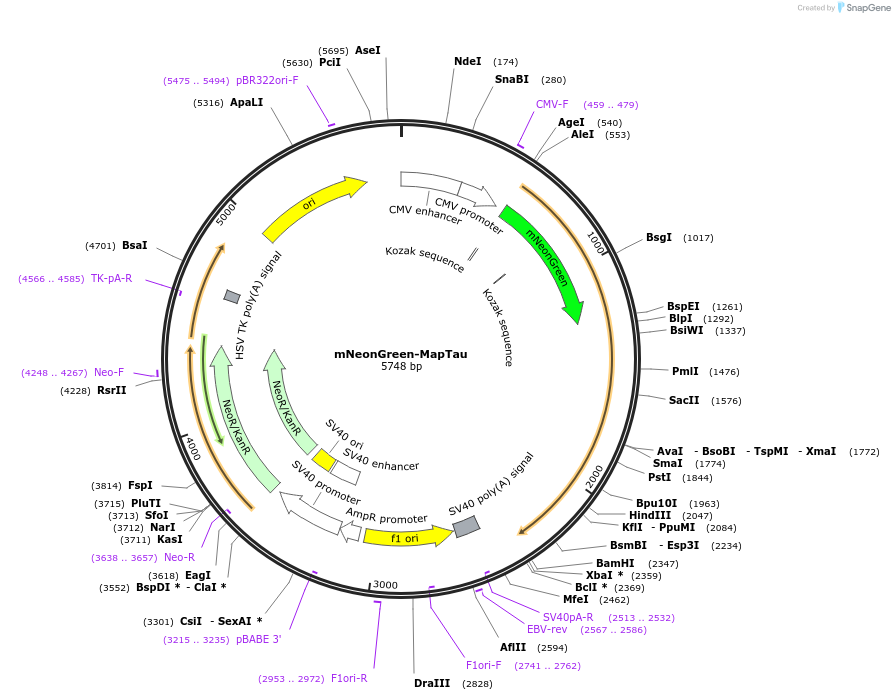
mNeonGreen-MapTau
Plasmid#137808PurposeVisualization of microtubulesDepositorInsertMapTau
ExpressionMammalianPromoterCMVAvailable SinceDec. 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
pDRF1 mScarletI3-T2A-mTurquoise2
Plasmid#219845Purposeequimolar expression of mScarletI3 and mTurquoise2 in budding yeastDepositorInsertmScarletI3-T2A-mTurquoise2
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
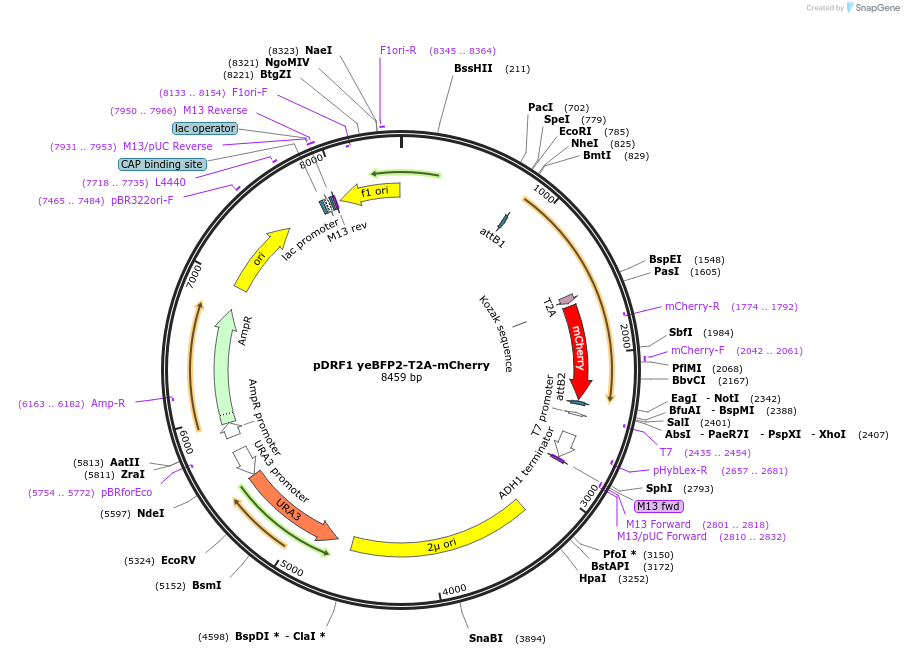
pDRF1 yeBFP2-T2A-mCherry
Plasmid#219851Purposeequimolar expression of yeBFP2 and mCherry in budding yeastDepositorInsertyeBFP2-T2A-mCherry
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
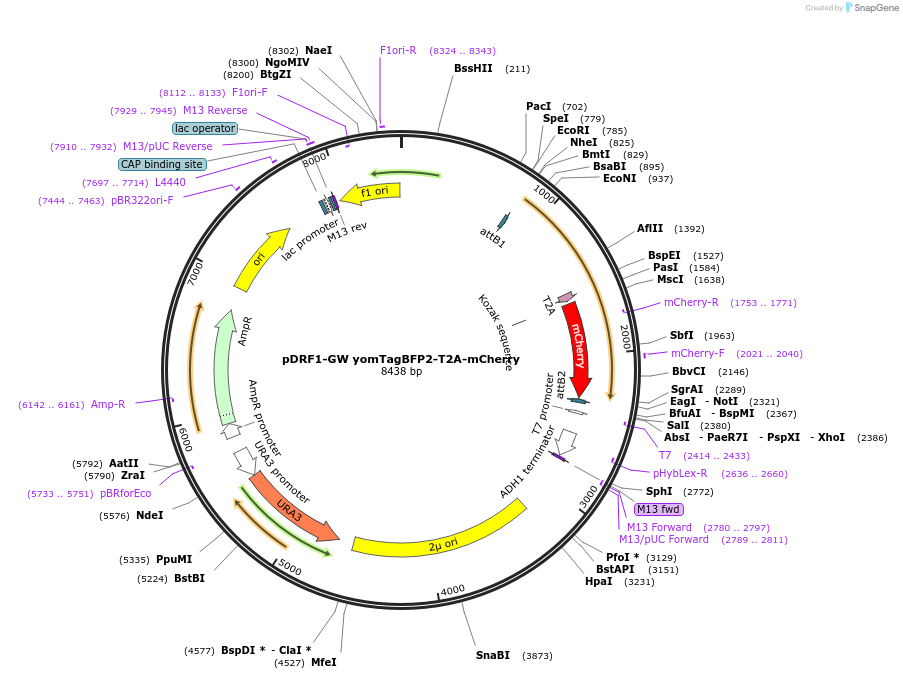
pDRF1-GW yomTagBFP2-T2A-mCherry
Plasmid#205493PurposeExpression of yomTagBFP2-T2A-mCherry in yeastDepositorInsertyomTagBFP2-T2A-mCherry
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
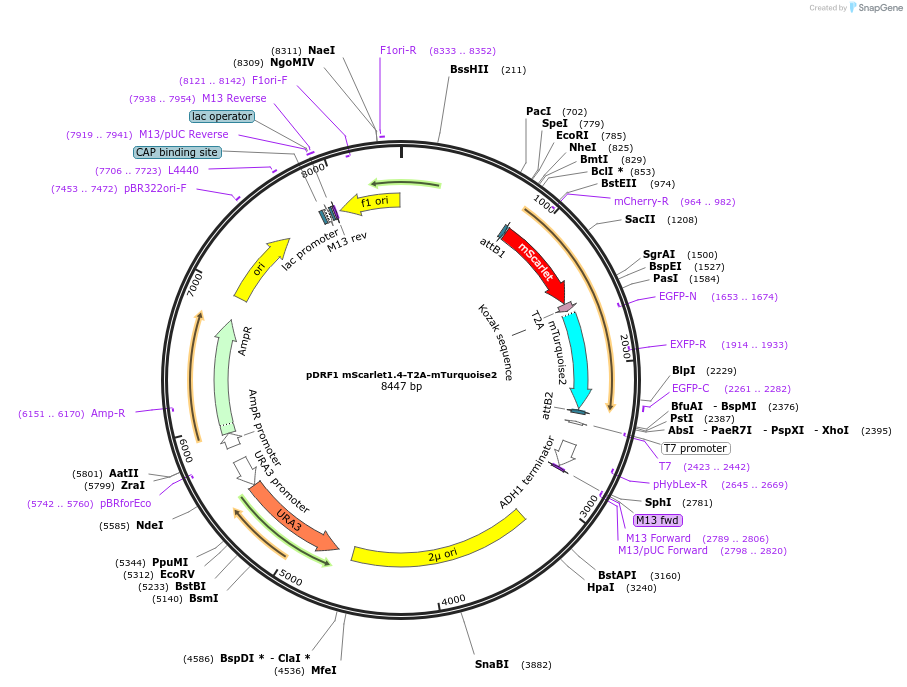
pDRF1 mScarlet1.4-T2A-mTurquoise2
Plasmid#219849Purposeequimolar expression of mScarlet1.4 and mTurquoise2 in budding yeastDepositorInsertmScarlet1.4-T2A-mTurquoise2
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
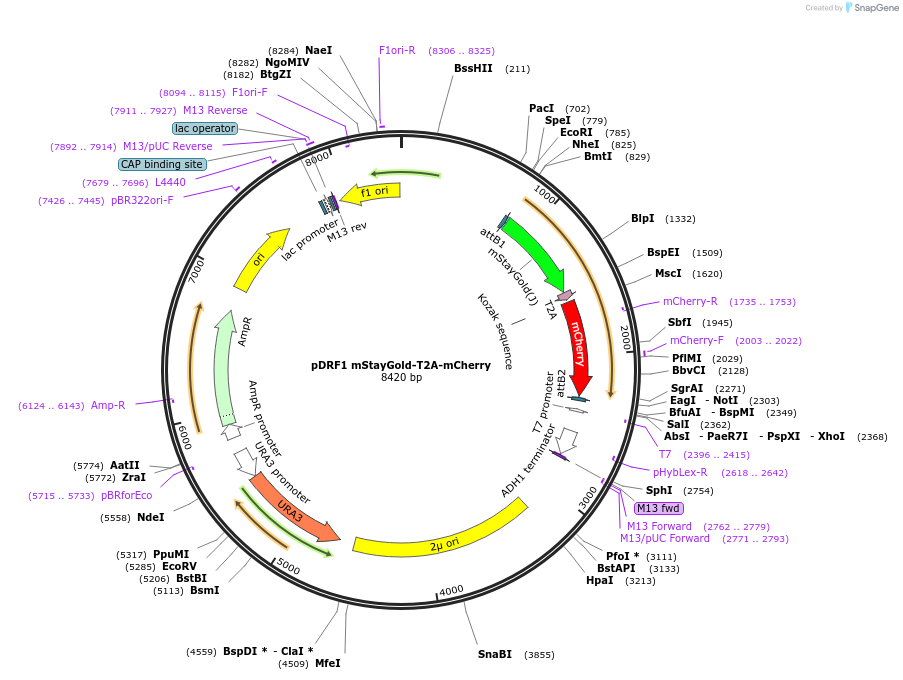
pDRF1 mStayGold-T2A-mCherry
Plasmid#219834Purposeequimolar expression of mStaygold and mCherry in budding yeastDepositorInsertmStayGold-T2A-mCherry
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
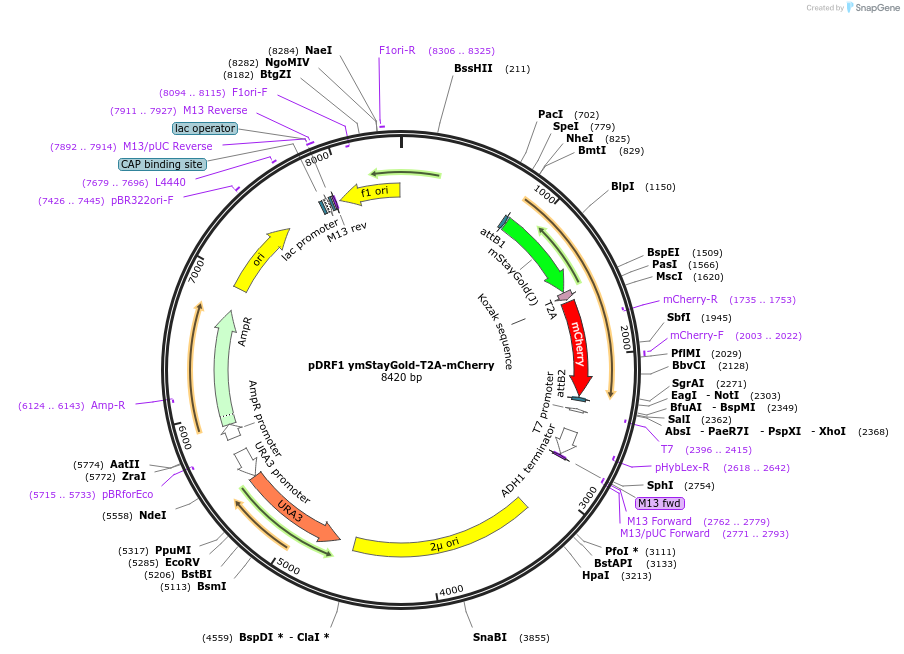
pDRF1 ymStayGold-T2A-mCherry
Plasmid#219843Purposeequimolar expression of ymStayGold and mCherry in budding yeastDepositorInsertymStayGold-T2A-mCherry
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
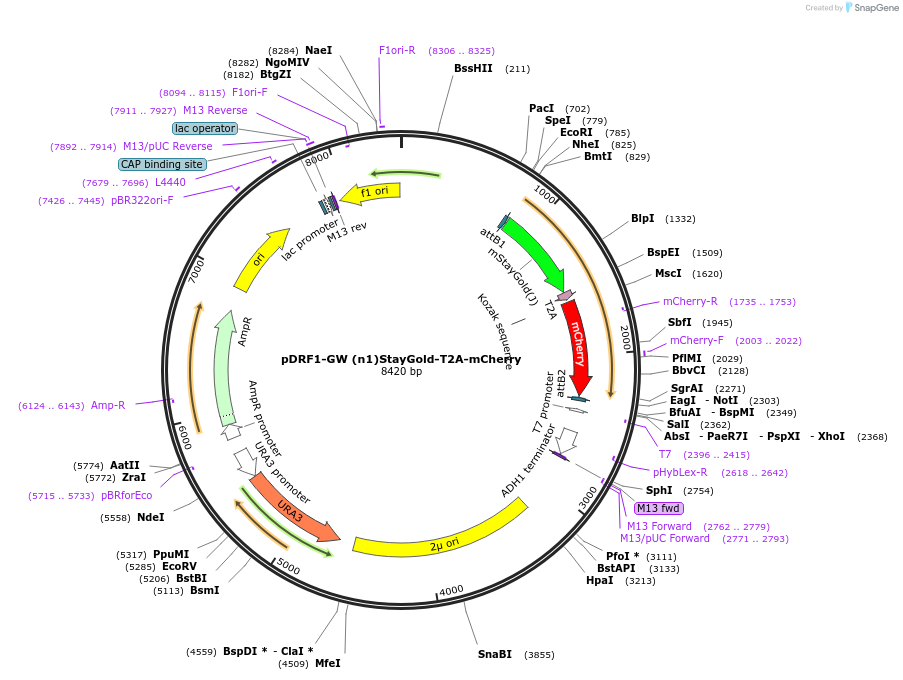
pDRF1-GW (n1)StayGold-T2A-mCherry
Plasmid#205495PurposeExpression of (n1)StayGold-T2A-mCherry in yeastDepositorInsert(n1)StayGold-T2A-mCherry
ExpressionYeastAvailable SinceSept. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
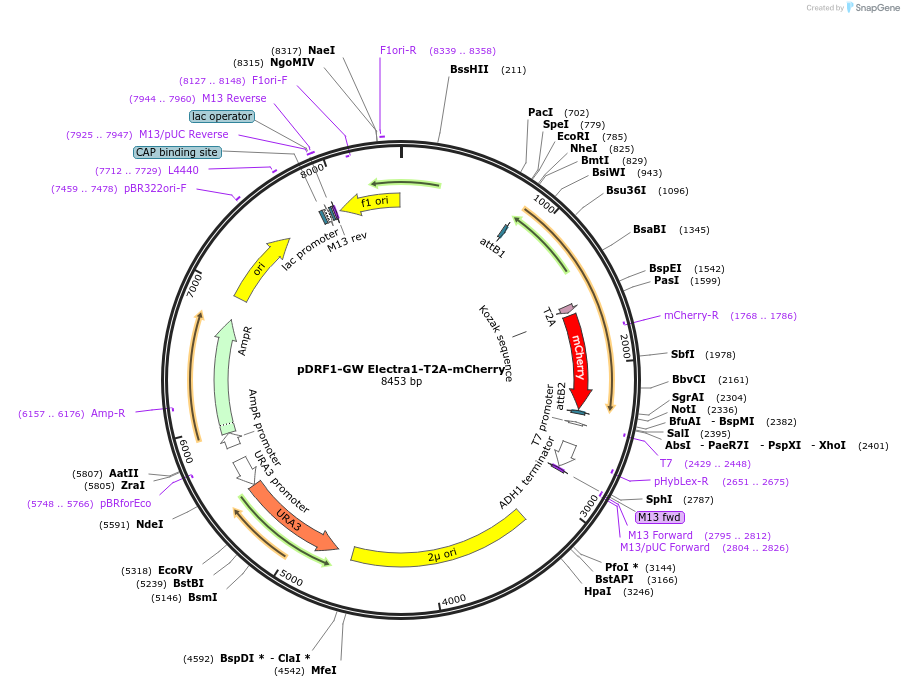
pDRF1-GW Electra1-T2A-mCherry
Plasmid#205498PurposeExpression of Electra1-T2A-mCherry in yeastDepositorInsertElectra1-T2A-mCherry
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
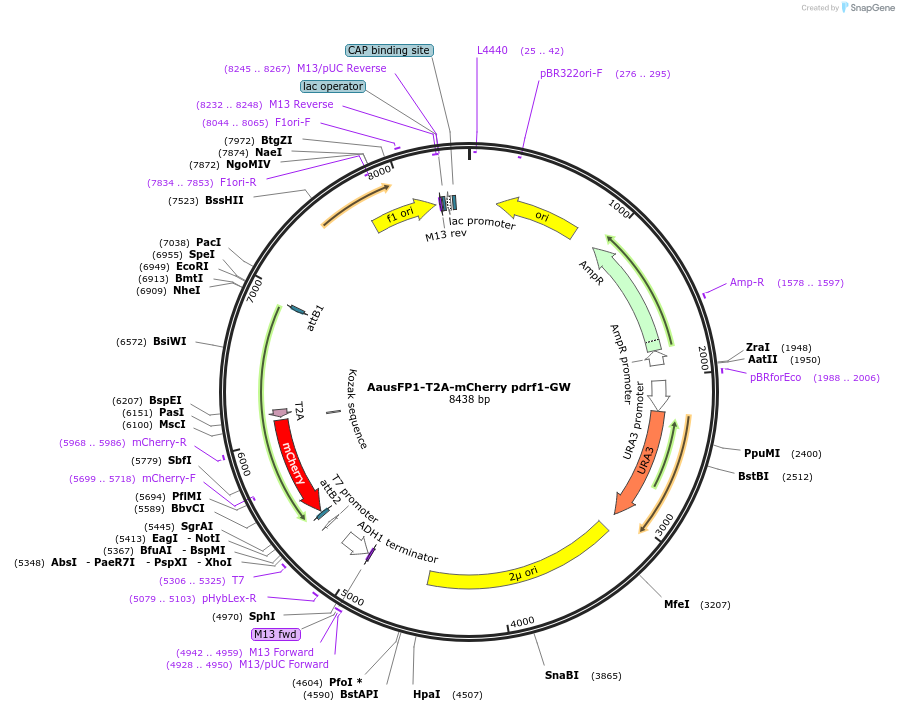
AausFP1-T2A-mCherry pdrf1-GW
Plasmid#168071PurposeProduces equimolar expression of AausFP1 and mCherry for use in yeastDepositorInsertAausFP1-T2A-mCherry
ExpressionYeastAvailable SinceJune 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
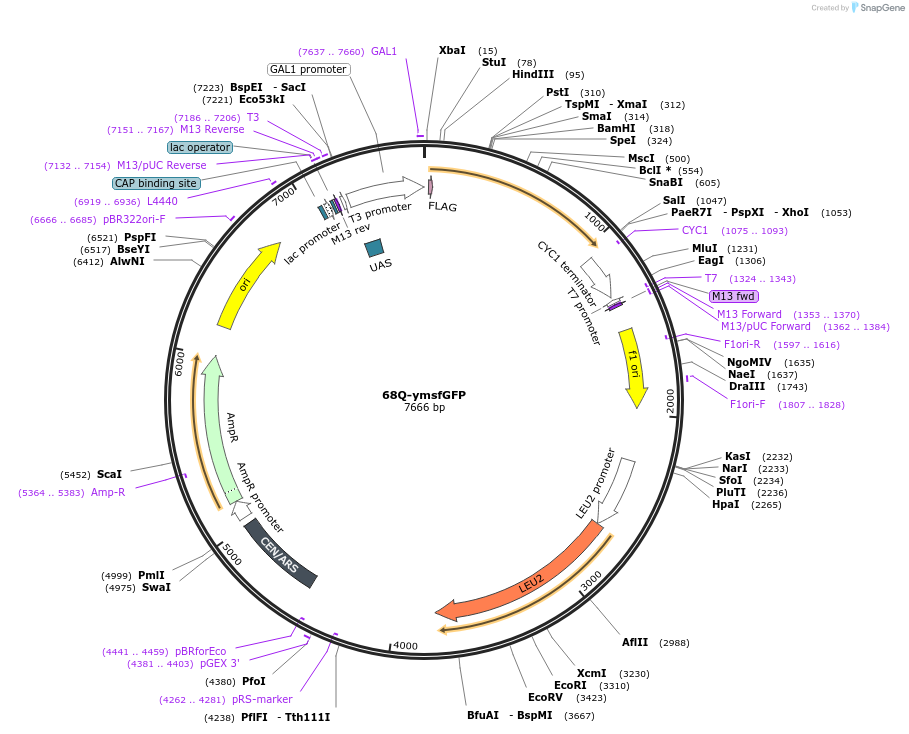
68Q-ymsfGFP
Plasmid#128560PurposeHttex1 (without proline-rich region) fused to ymsfGFPDepositorInsert68Q-Httex1 (without proline-rich region) fused to ymsfGFP
TagsymsfGFPExpressionYeastPromoterGAL1Available SinceOct. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
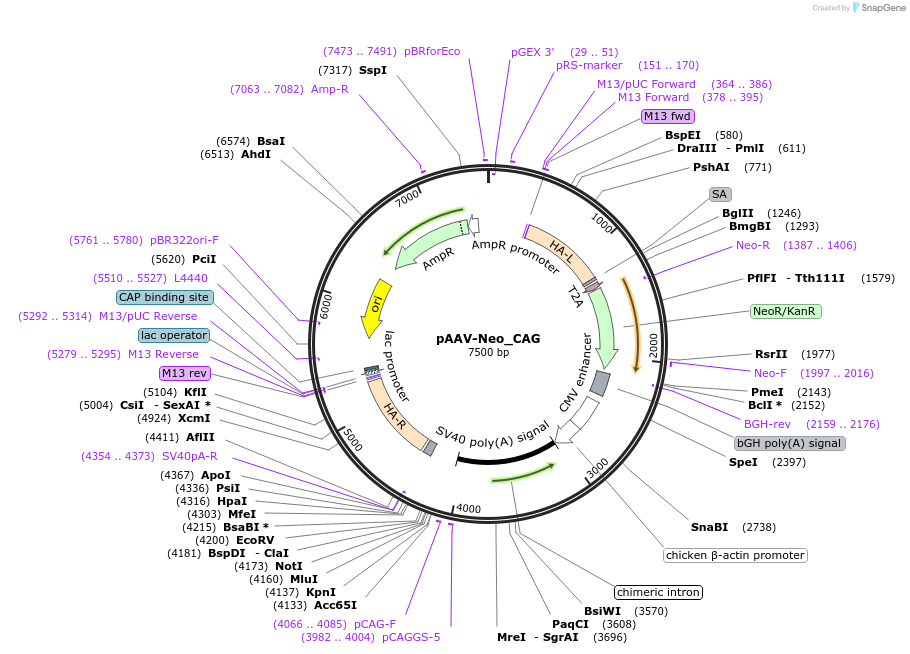
pAAV-Neo_CAG
Plasmid#220537PurposeAAVS1-targeting backbone used as a filler plasmid to co-deliver during nucleofection with pAAV-Puro_siKD2.0 (Plasmid #220536) cloned with shRNAsDepositorTypeEmpty backboneExpressionBacterialAvailable SinceDec. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
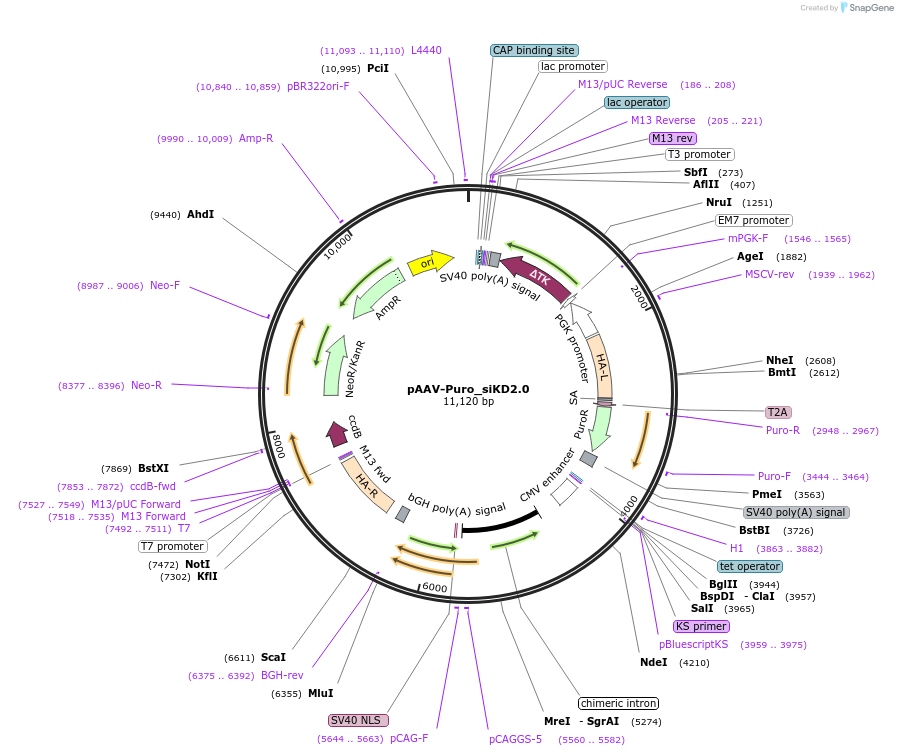
pAAV-Puro_siKD2.0
Plasmid#220536PurposeAAVS1-targeting vector designed for the iPS2-seq platform to express a single-copy, tetracycline-inducible shRNA in human pluripotent stem cellsDepositorTypeEmpty backboneExpressionBacterialAvailable SinceDec. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
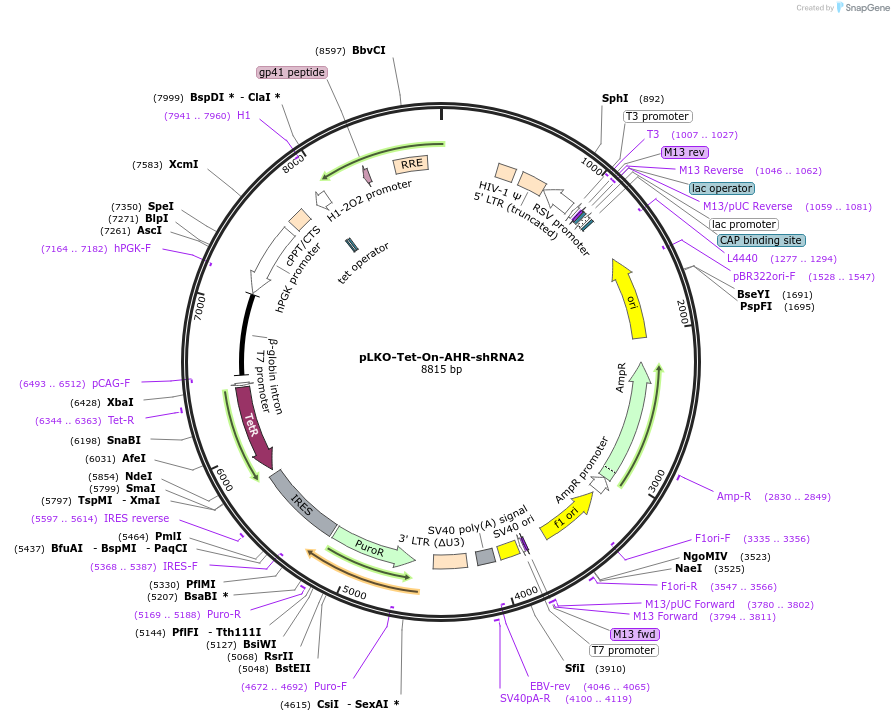
pLKO-Tet-On-AHR-shRNA2
Plasmid#166908PurposeLentivirus for inducible knockdown of human AHRDepositorInsertAHR shRNA
UseLentiviralExpressionMammalianAvailable SinceOct. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
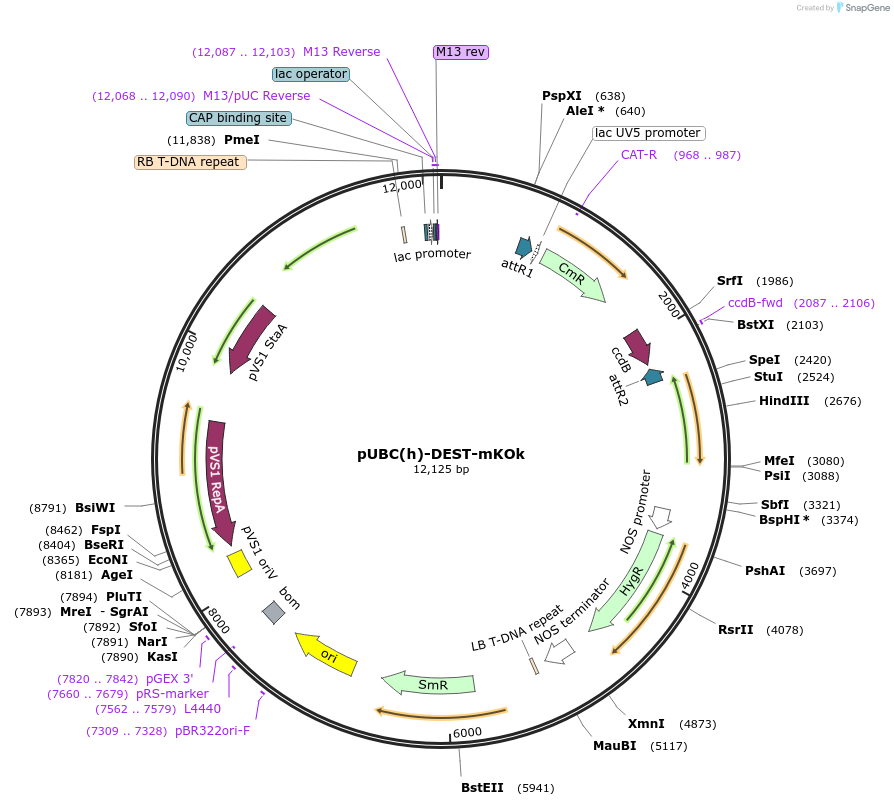
pUBC(h)-DEST-mKOk
Plasmid#224830PurposeEmpty Gateway DEST vector for expressing C-terminal tagged proteins with mKusabiraOrangekand hygromycin selection in plantsDepositorTypeEmpty backboneTagsmKusabiraOrangekExpressionPlantAvailable SinceJuly 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
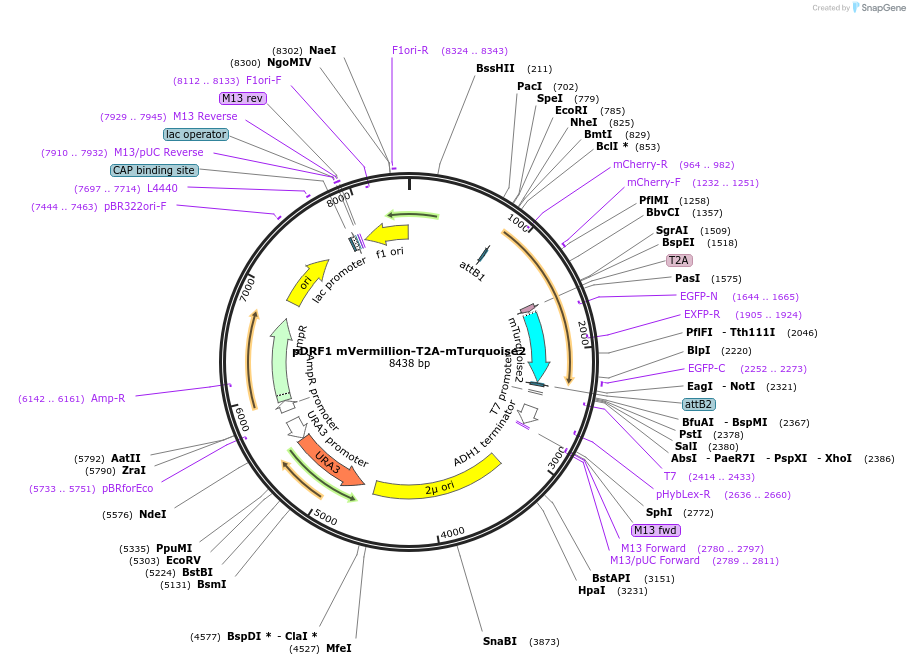
pDRF1 mVermillion-T2A-mTurquoise2
Plasmid#219847Purposeequimolar expression of mVermillion and mTurquoise2 in budding yeastDepositorInsertmVermillion-T2A-mTurquoise2
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
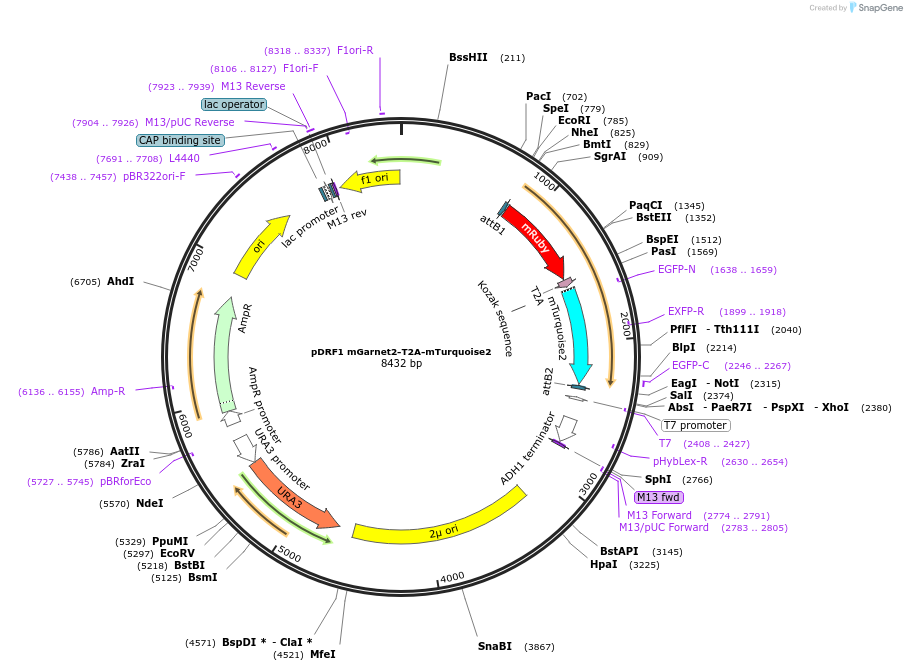
pDRF1 mGarnet2-T2A-mTurquoise2
Plasmid#219850Purposeequimolar expression of mGarnet2 and mTurquoise2 in budding yeastDepositorInsertmGarnet2-T2A-mTurquoise2
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
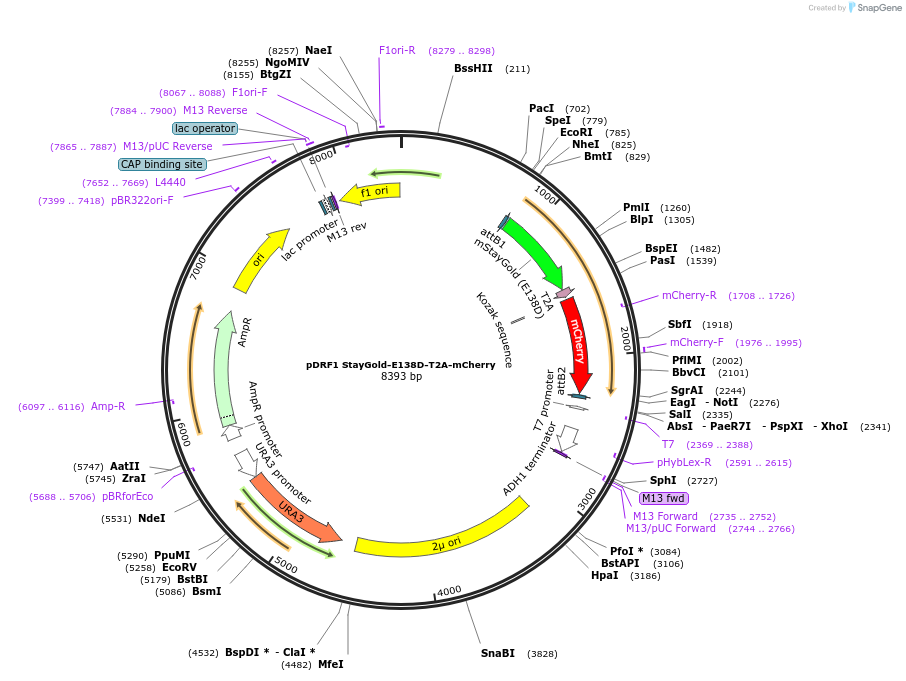
pDRF1 StayGold-E138D-T2A-mCherry
Plasmid#219833Purposeequimolar expression of Staygold-E138D and mCherry in budding yeastDepositorInsertStayGold-E138D-T2A-mCherry
ExpressionYeastAvailable SinceAug. 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
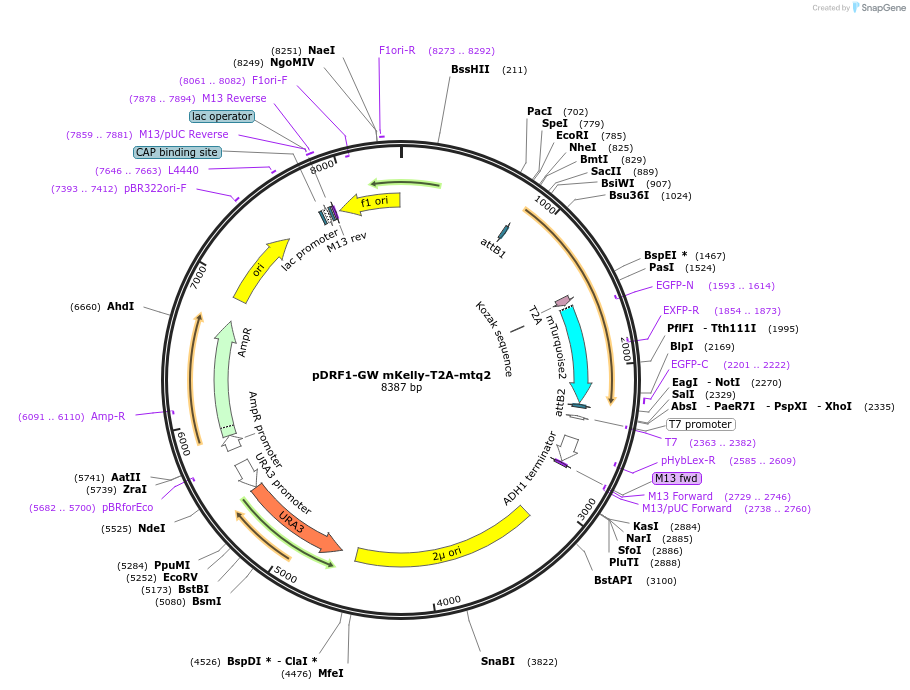
pDRF1-GW mKelly-T2A-mtq2
Plasmid#205506PurposeExpression of mKelly-T2A-mtq2 in yeastDepositorInsertmKelly-T2A-mtq2
ExpressionYeastAvailable SinceJuly 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
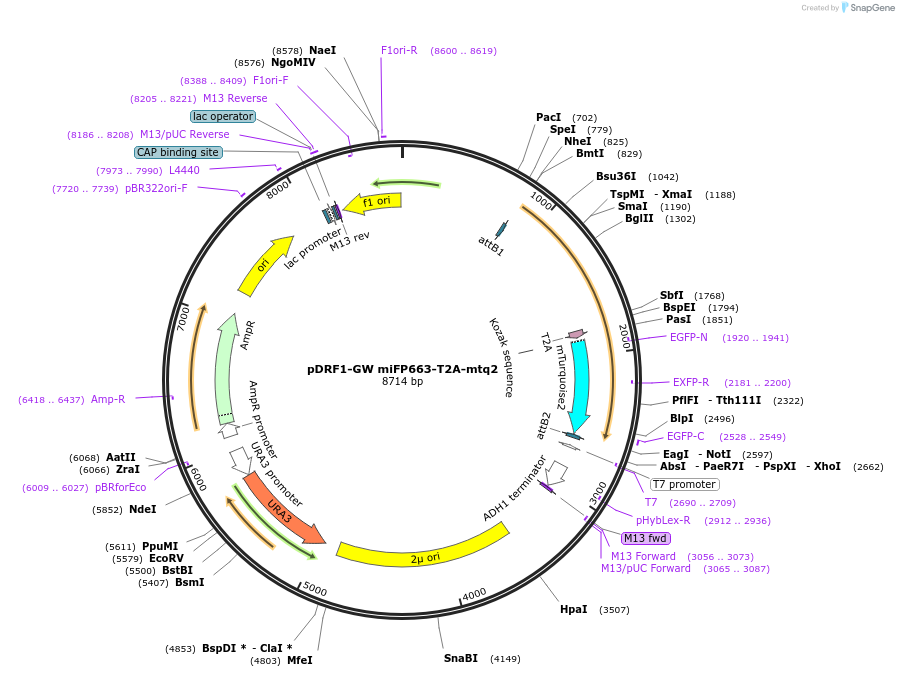
pDRF1-GW miFP663-T2A-mtq2
Plasmid#205504PurposeExpression of miFP663-T2A-mtq2 in yeastDepositorInsertmiFP663-T2A-mtq2
ExpressionYeastAvailable SinceSept. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
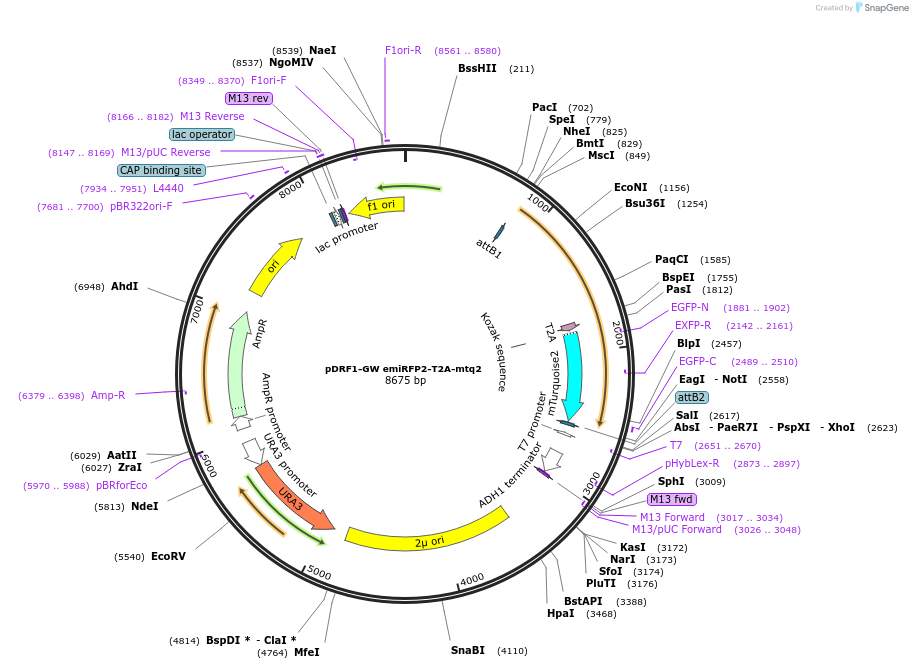
pDRF1-GW emiRFP2-T2A-mtq2
Plasmid#205497PurposeExpression of emiRFP2-T2A-mtq2 in yeastDepositorInsertemiRFP2-T2A-mtq2
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
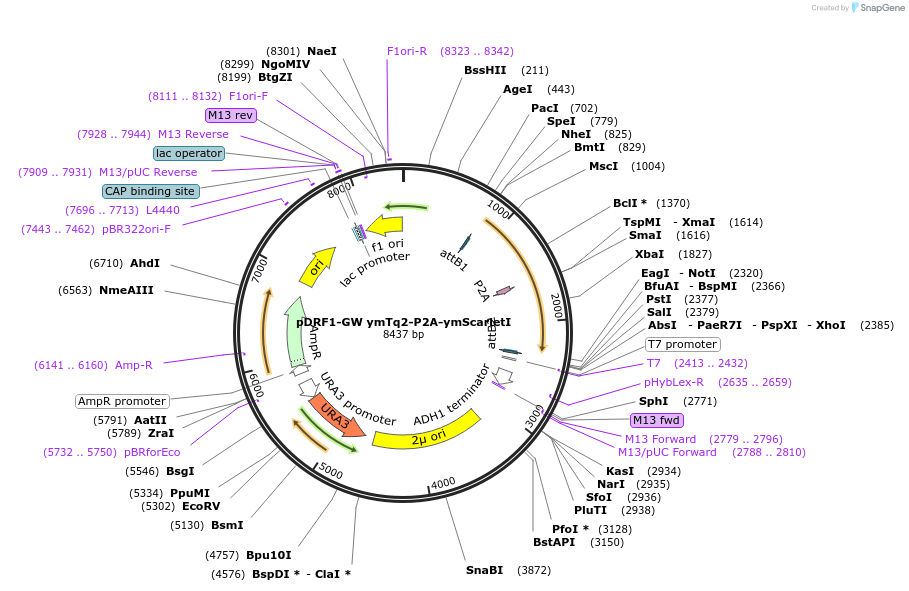
pDRF1-GW ymTq2-P2A-ymScarletI
Plasmid#205500PurposeExpression of ymTq2-P2A-ymScarletI in yeastDepositorInsertymTq2-P2A-ymScarletI
ExpressionYeastAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only