We narrowed to 14,493 results for: nts
-
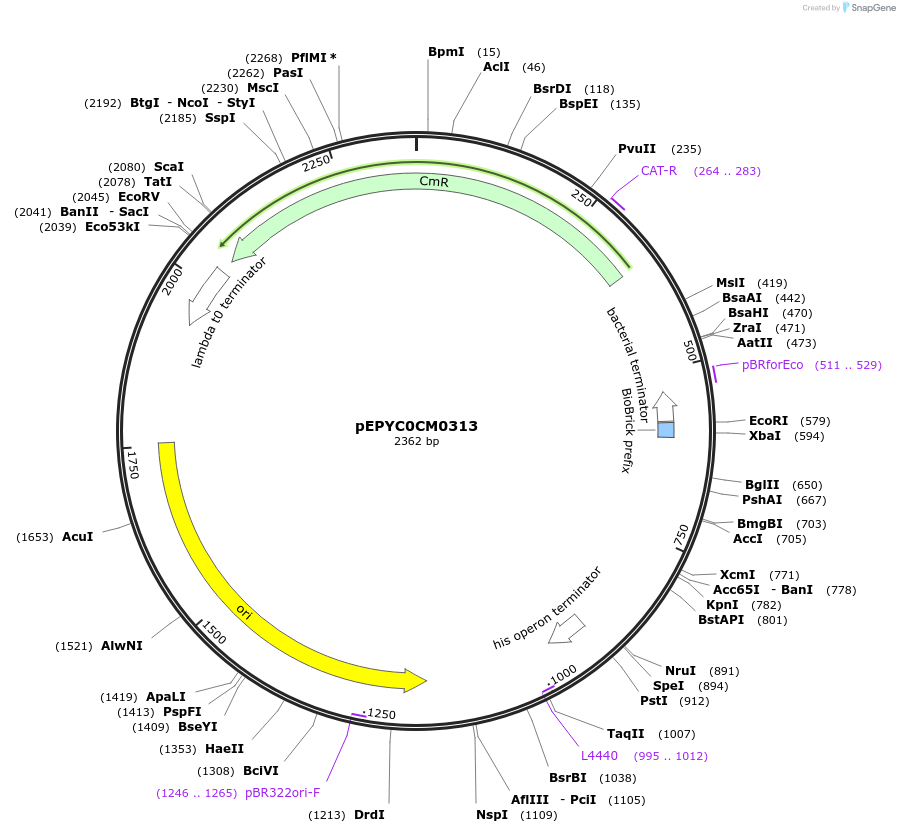
Plasmid#154538PurposePhytobricks Level 0 partDepositorInsertMinSyn_036
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
pEPYC0CM0314
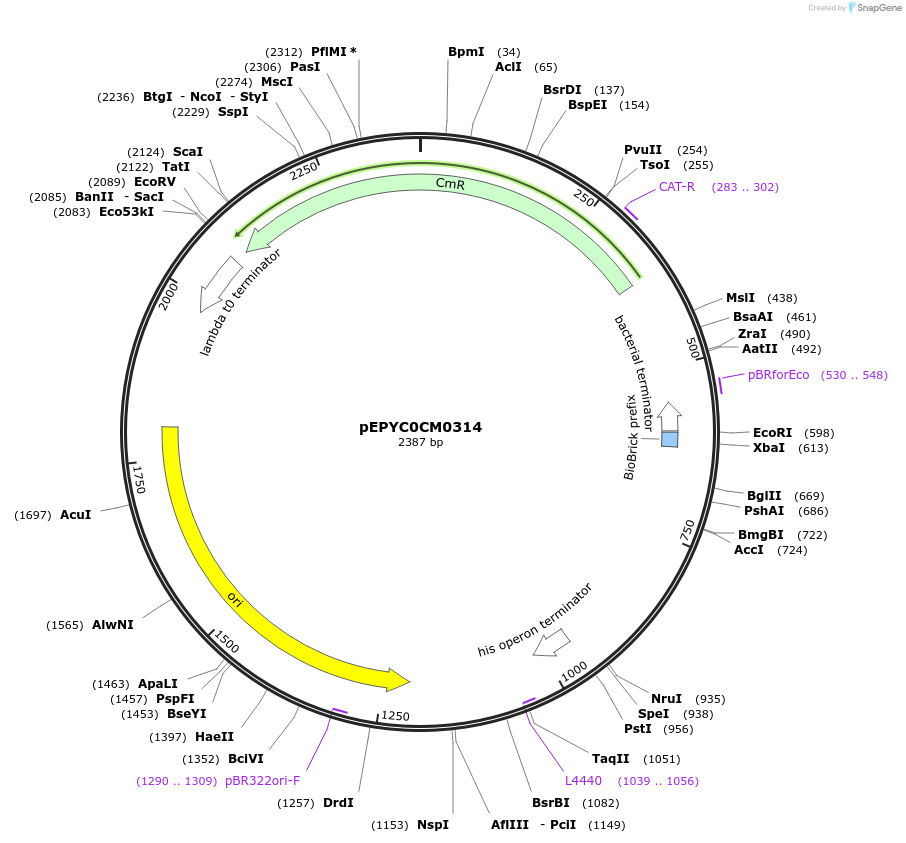
Plasmid#154539PurposePhytobricks Level 0 partDepositorInsertMinSyn_037
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
pEPYC0CM0225
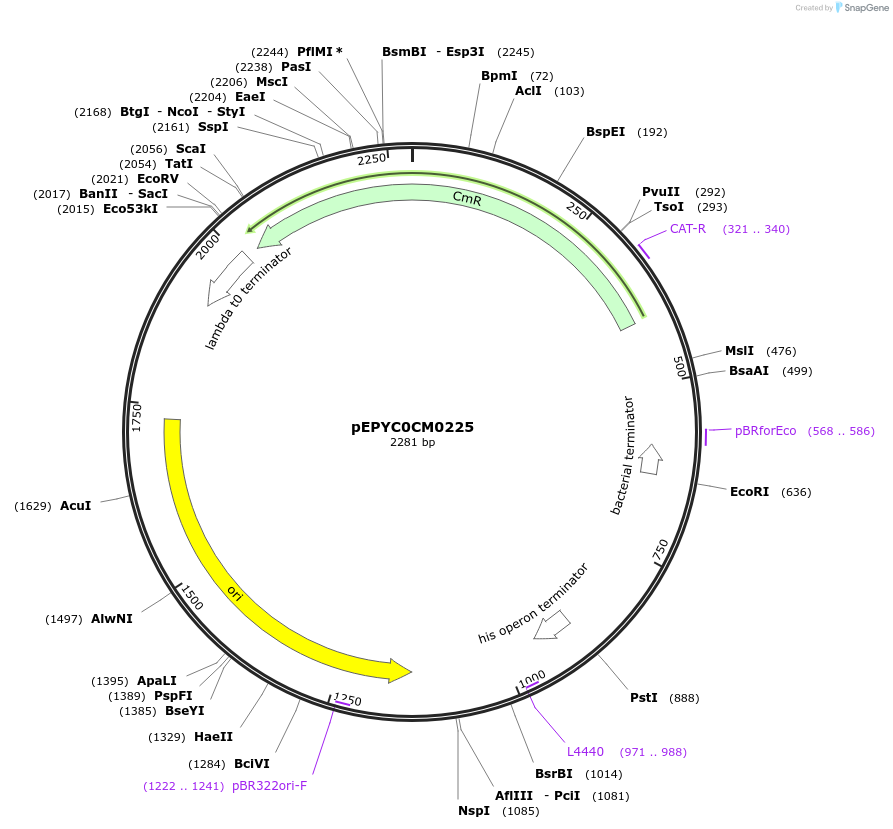
Plasmid#154540PurposePhytobricks Level 0 partDepositorInsertMinSyn_038
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
pEPYC0CM0209
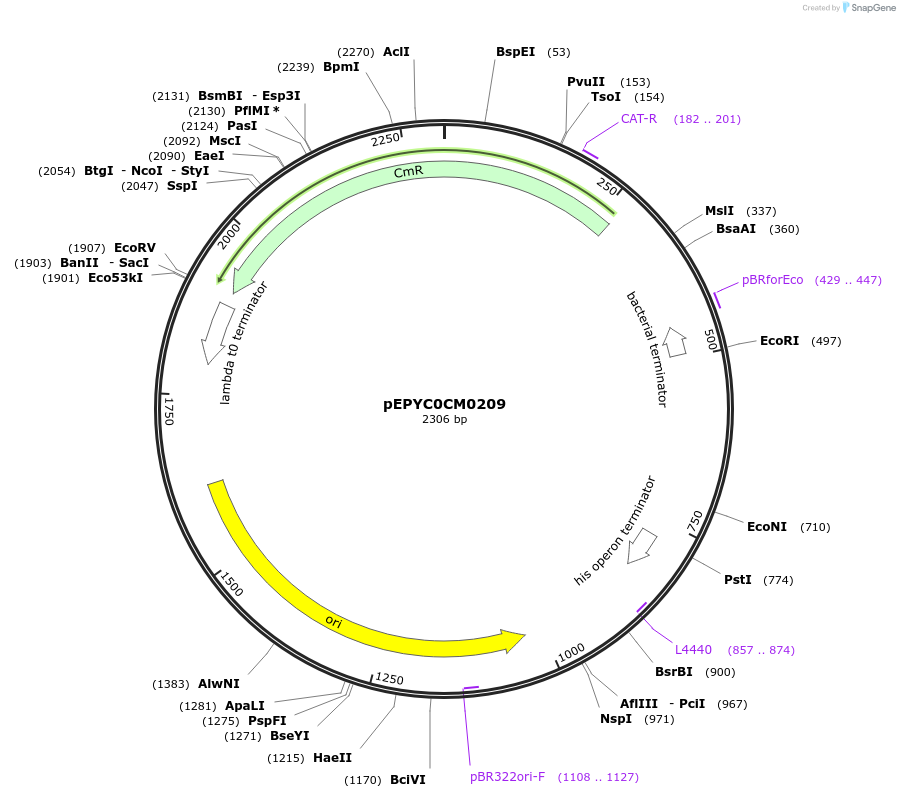
Plasmid#154541PurposePhytobricks Level 0 partDepositorInsertMinSyn_039
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
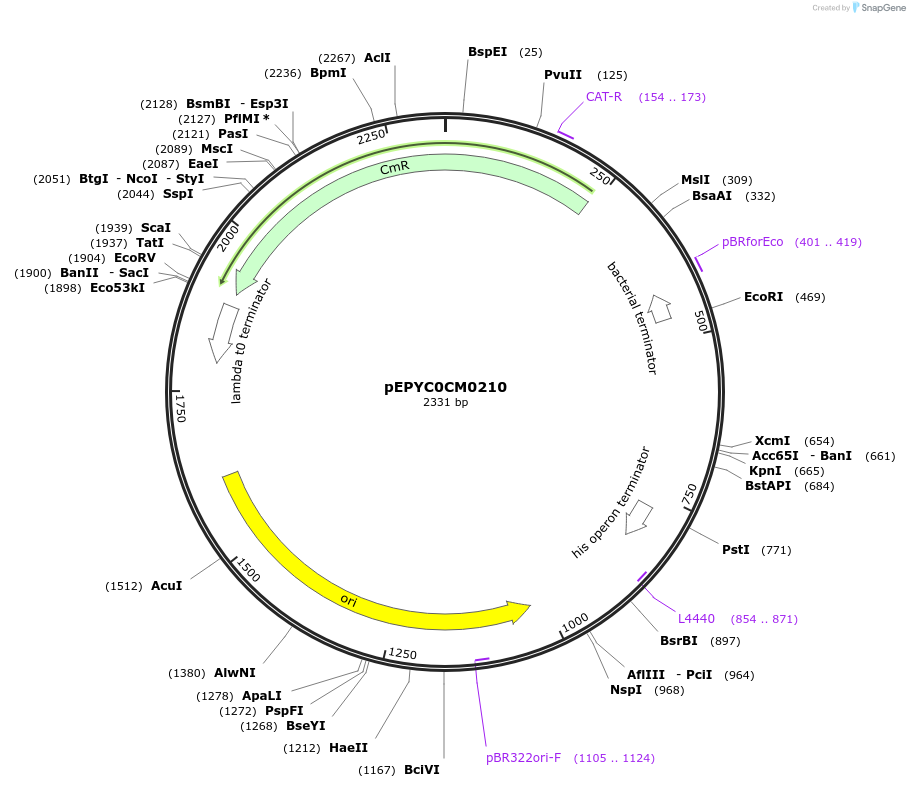
pEPYC0CM0210
Plasmid#154542PurposePhytobricks Level 0 partDepositorInsertMinSyn_040
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
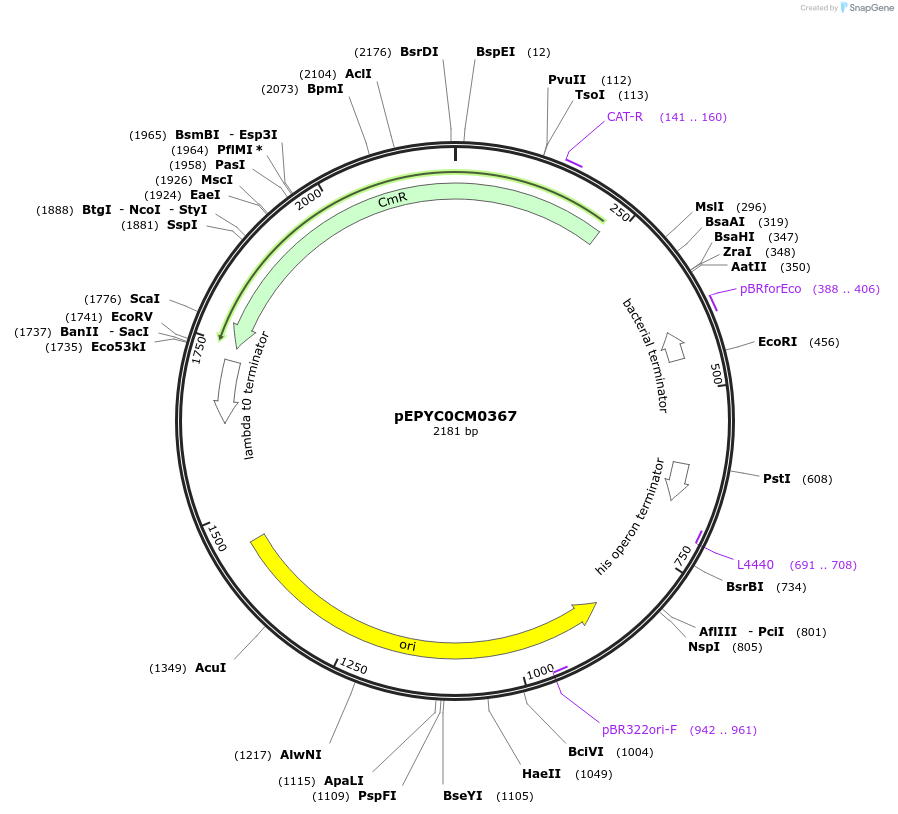
pEPYC0CM0367
Plasmid#154505PurposePhytobricks Level 0 partDepositorInsertMinSyn_002
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
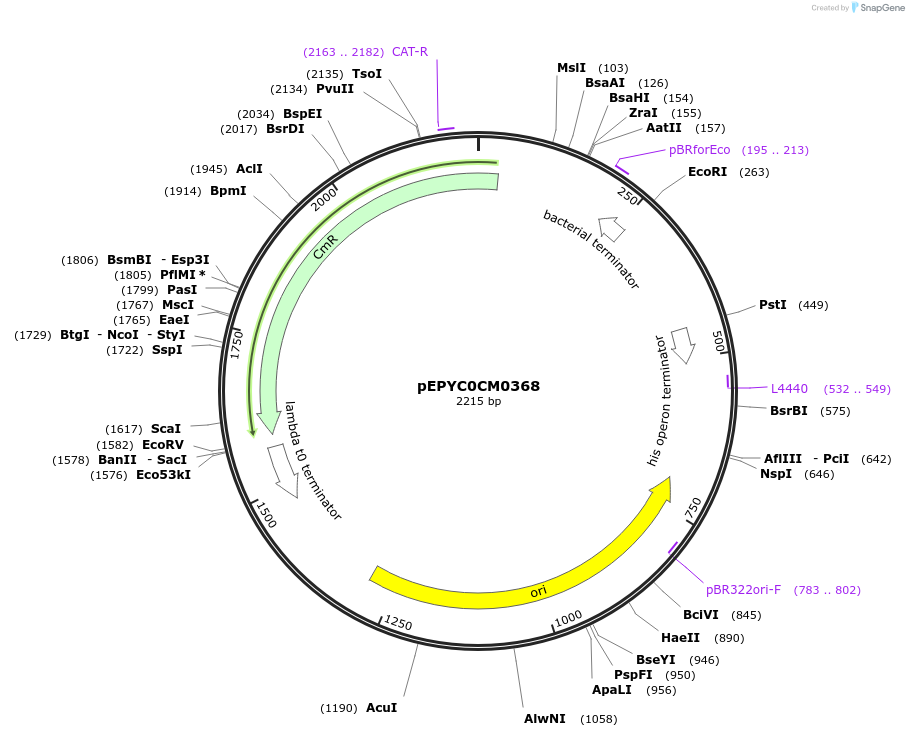
pEPYC0CM0368
Plasmid#154506PurposePhytobricks Level 0 partDepositorInsertMinSyn_003
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
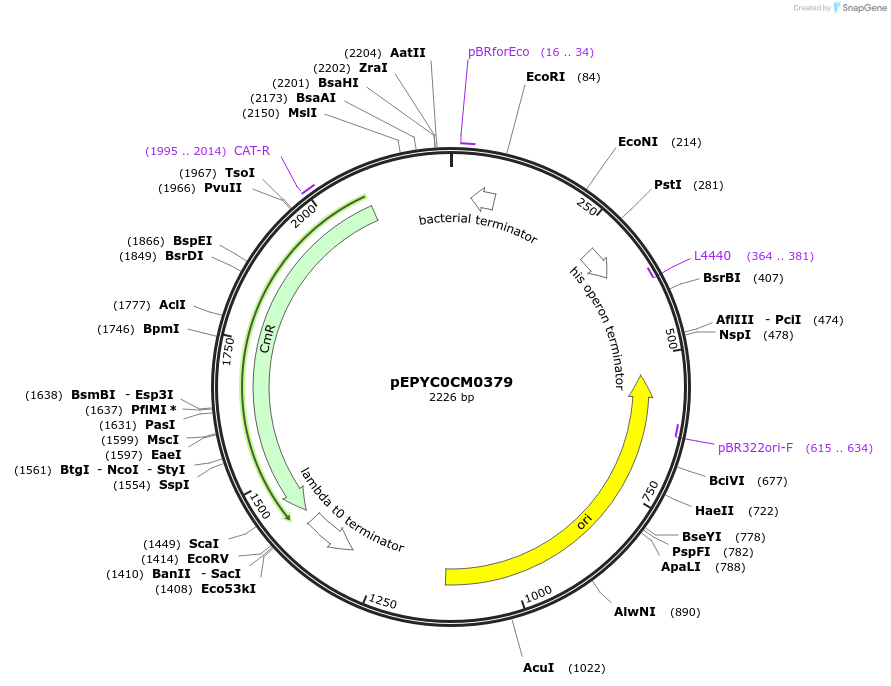
pEPYC0CM0379
Plasmid#154507PurposePhytobricks Level 0 partDepositorInsertMinSyn_004
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
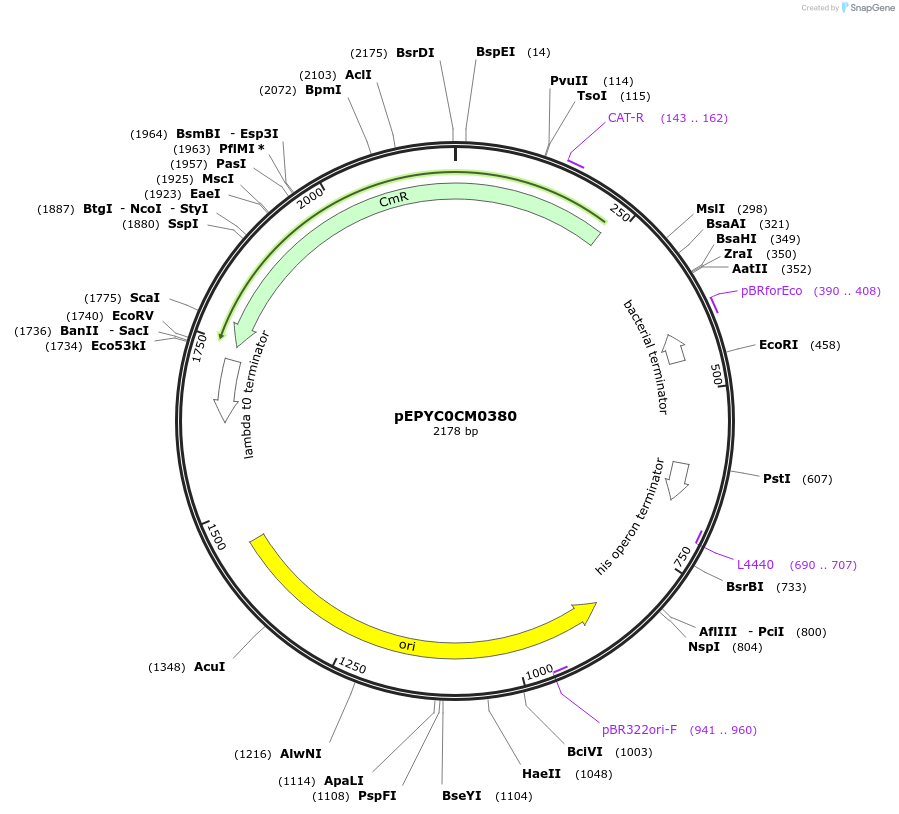
pEPYC0CM0380
Plasmid#154508PurposePhytobricks Level 0 partDepositorInsertMinSyn_005
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
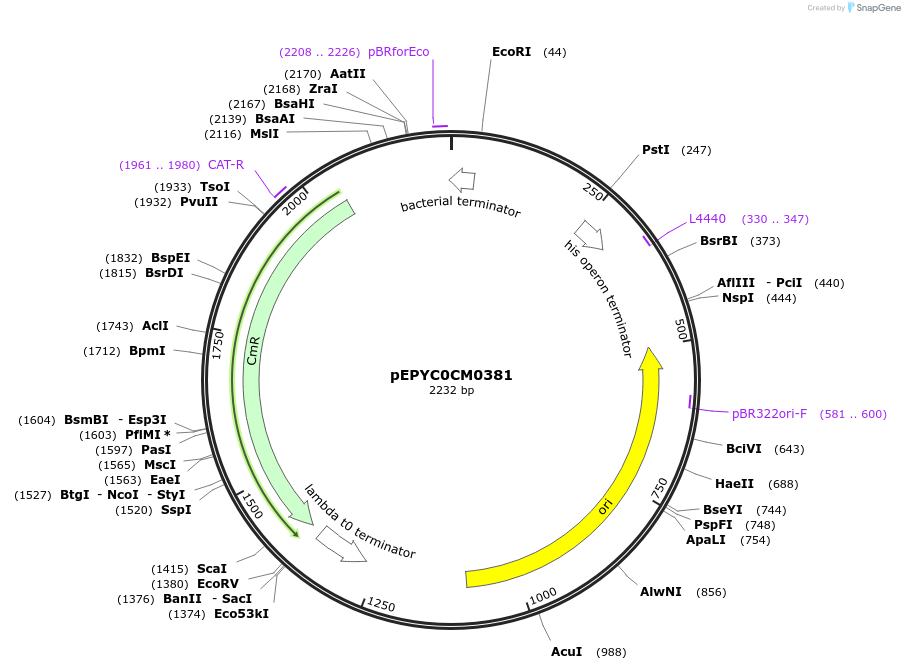
pEPYC0CM0381
Plasmid#154509PurposePhytobricks Level 0 partDepositorInsertMinSyn_006
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
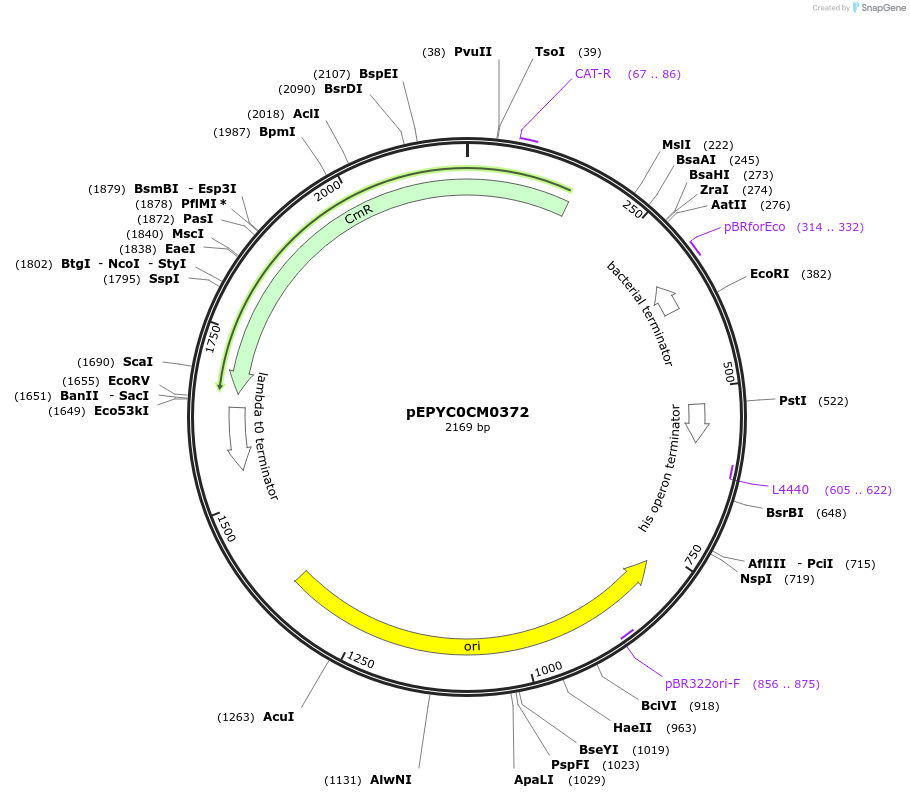
pEPYC0CM0372
Plasmid#154510PurposePhytobricks Level 0 partDepositorInsertMinSyn_007
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
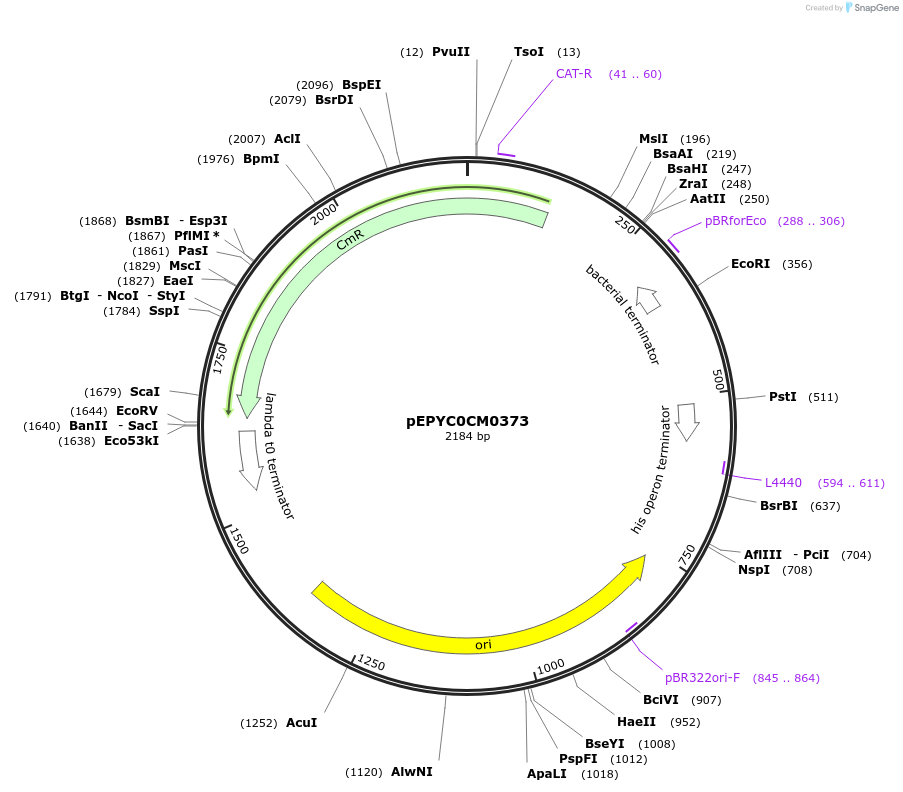
pEPYC0CM0373
Plasmid#154511PurposePhytobricks Level 0 partDepositorInsertMinSyn_008
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
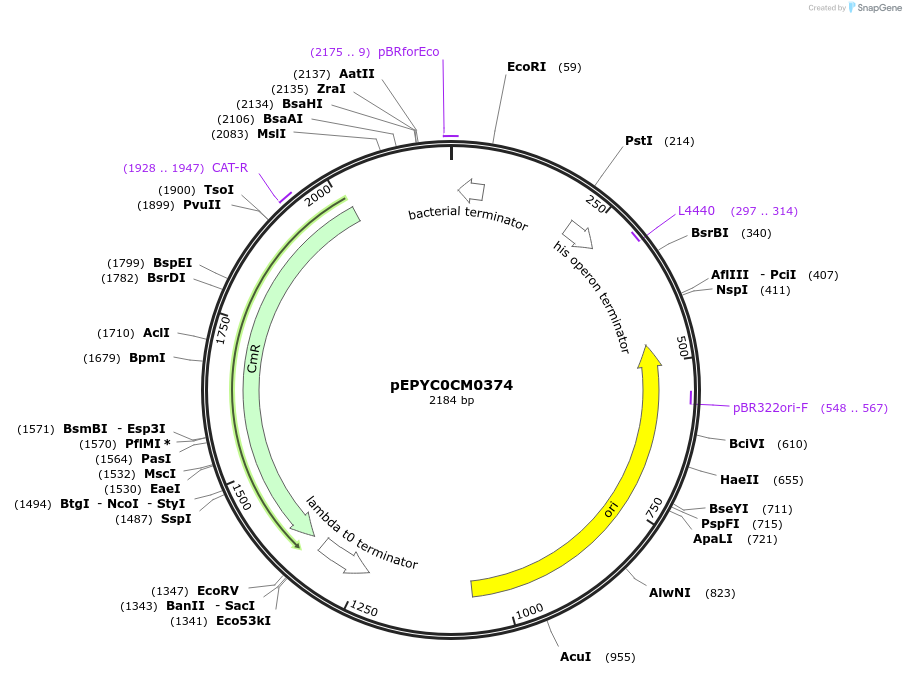
pEPYC0CM0374
Plasmid#154512PurposePhytobricks Level 0 partDepositorInsertMinSyn_009
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
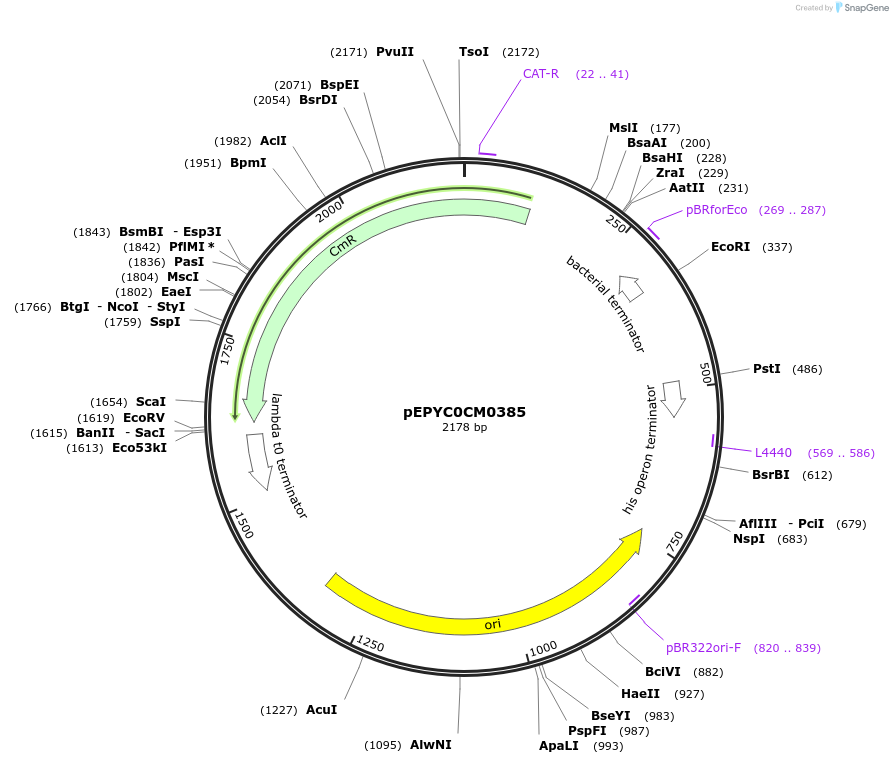
pEPYC0CM0385
Plasmid#154513PurposePhytobricks Level 0 partDepositorInsertMinSyn_010
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
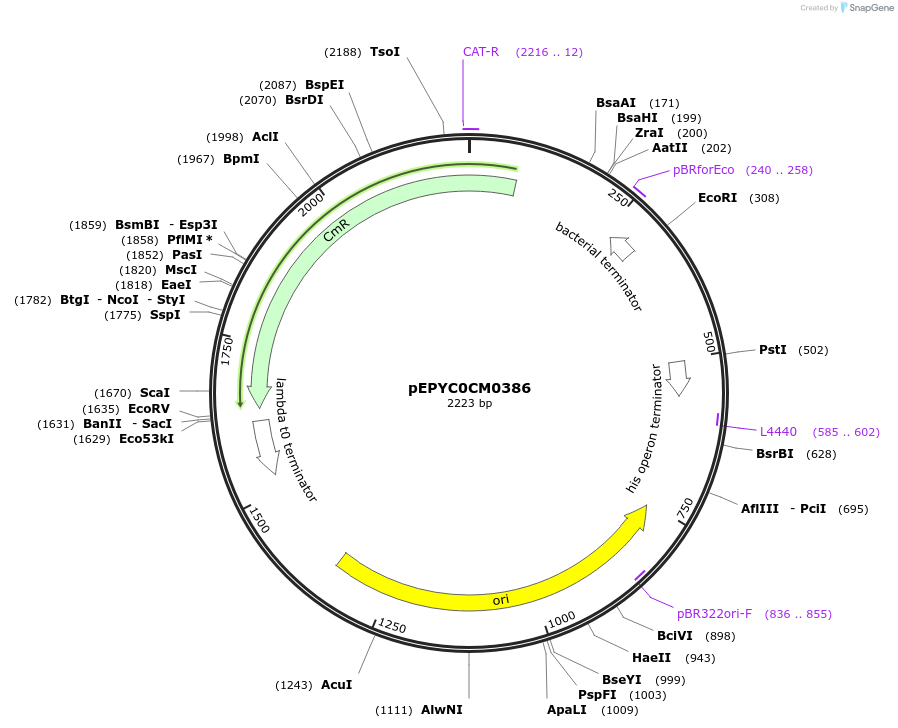
pEPYC0CM0386
Plasmid#154514PurposePhytobricks Level 0 partDepositorInsertMinSyn_011
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
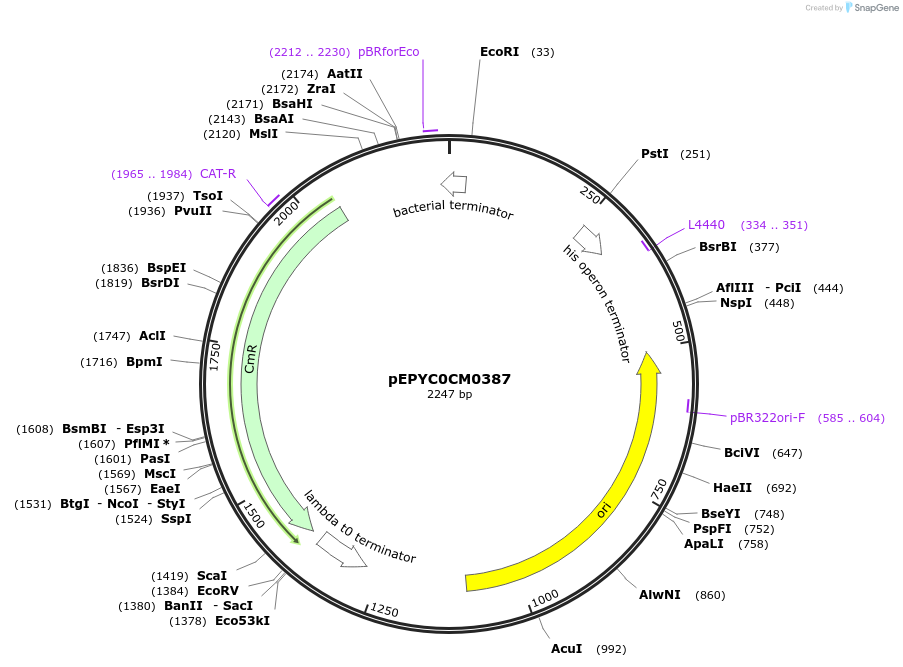
pEPYC0CM0387
Plasmid#154515PurposePhytobricks Level 0 partDepositorInsertMinSyn_012
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
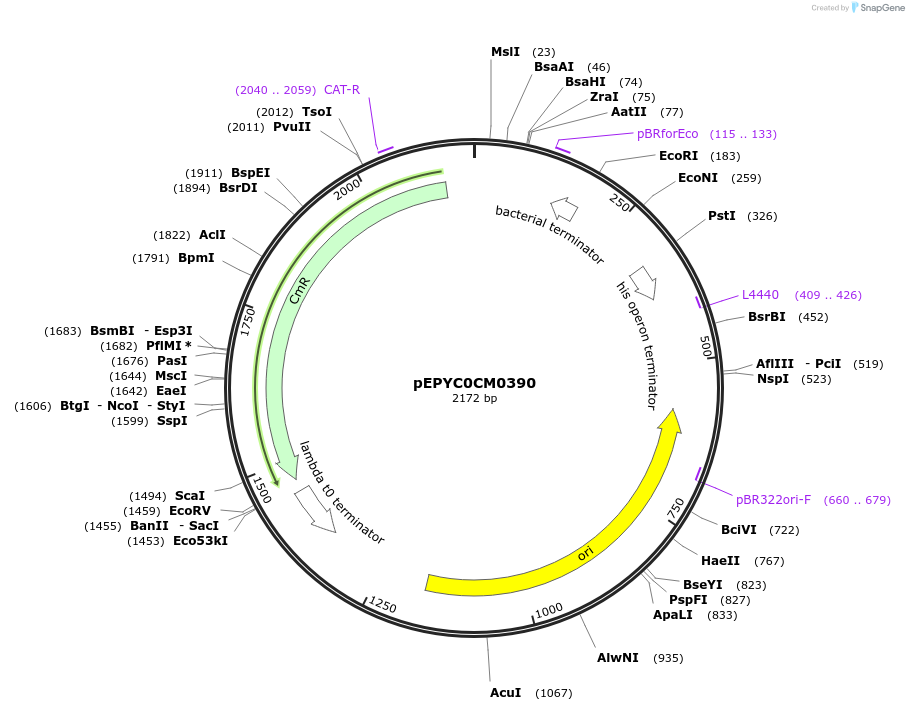
pEPYC0CM0390
Plasmid#154516PurposePhytobricks Level 0 partDepositorInsertMinSyn_013
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
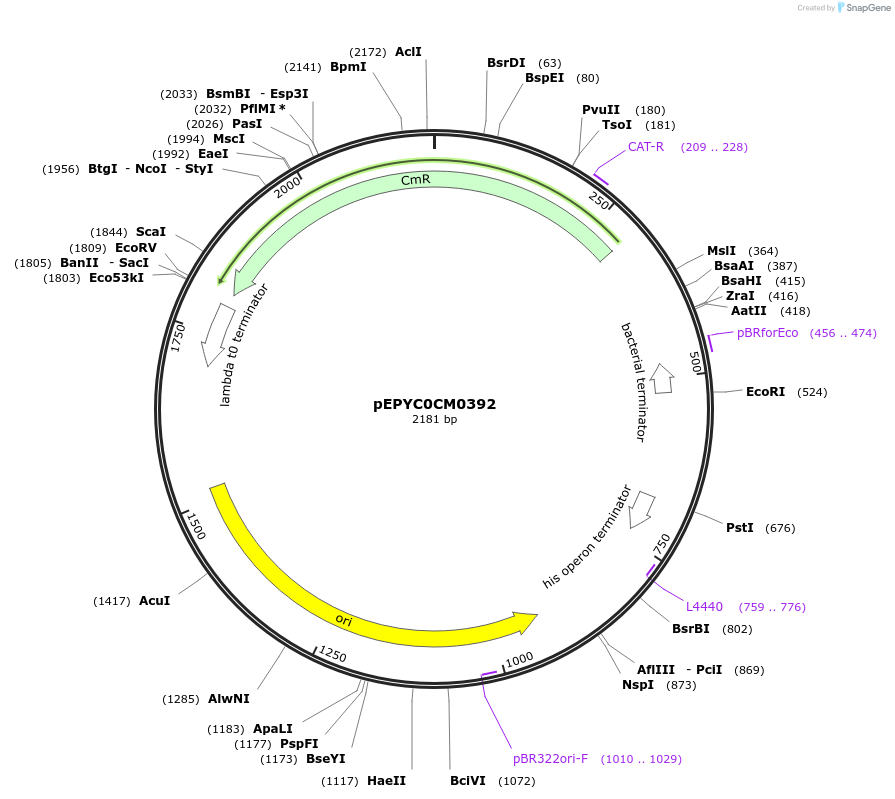
pEPYC0CM0392
Plasmid#154518PurposePhytobricks Level 0 partDepositorInsertMinSyn_015
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
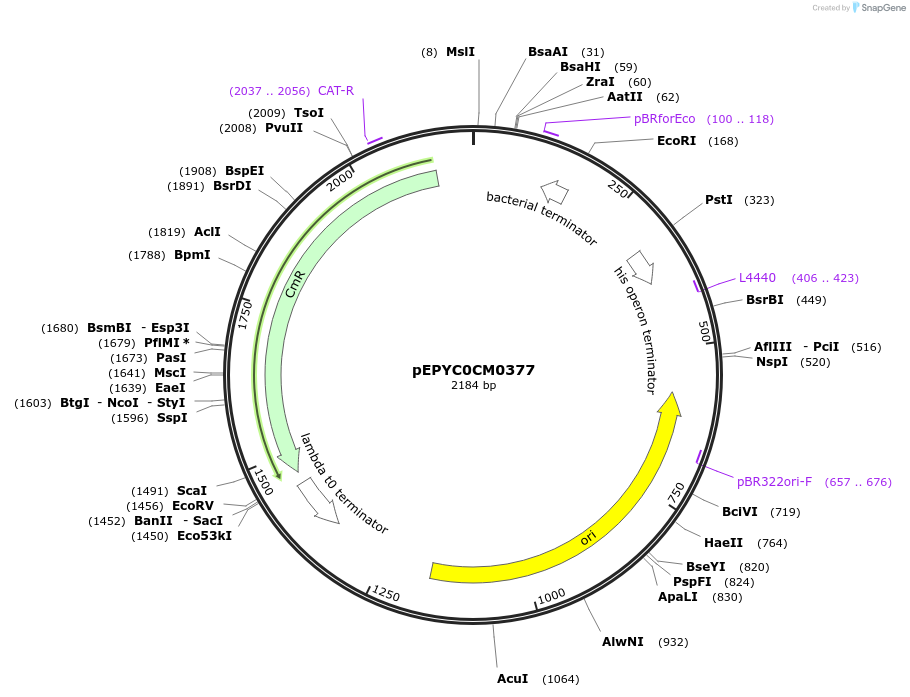
pEPYC0CM0377
Plasmid#154521PurposePhytobricks Level 0 partDepositorInsertMinSyn_018
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
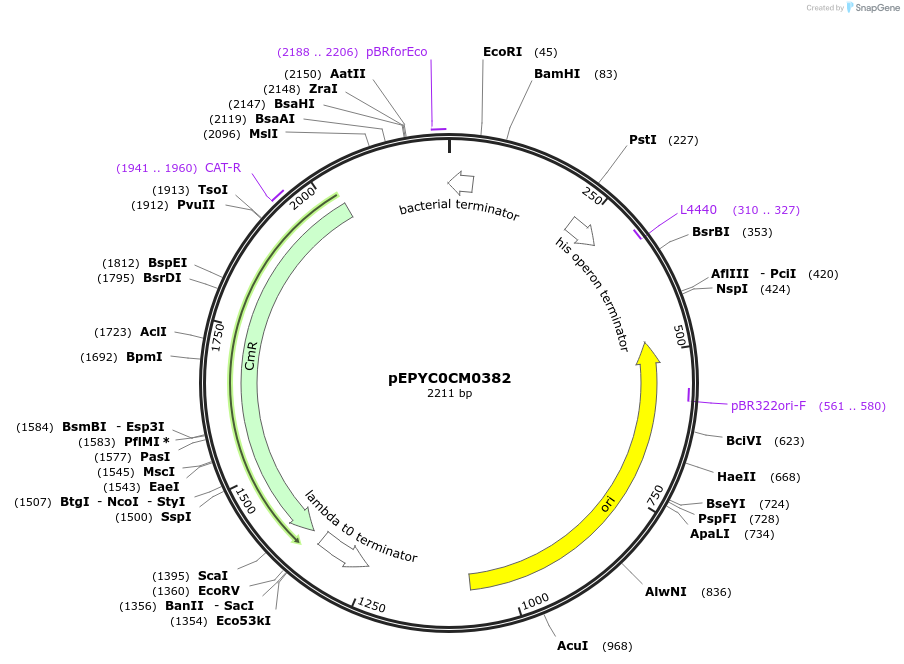
pEPYC0CM0382
Plasmid#154522PurposePhytobricks Level 0 partDepositorInsertMinSyn_019
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
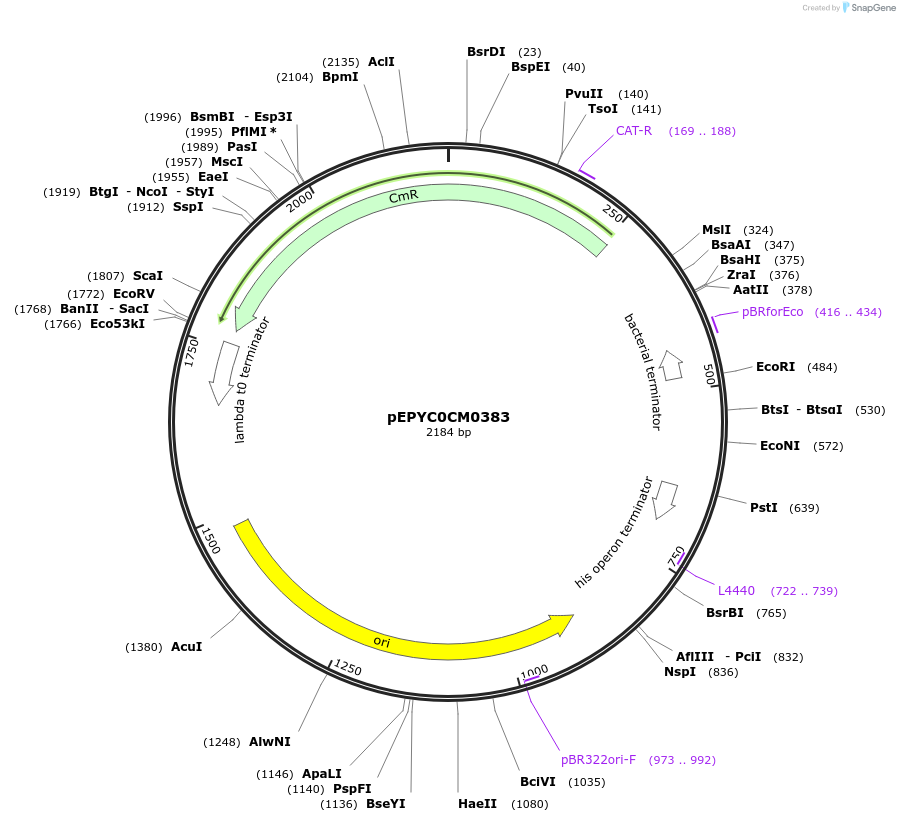
pEPYC0CM0383
Plasmid#154523PurposePhytobricks Level 0 partDepositorInsertMinSyn_020
UseSynthetic BiologyAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
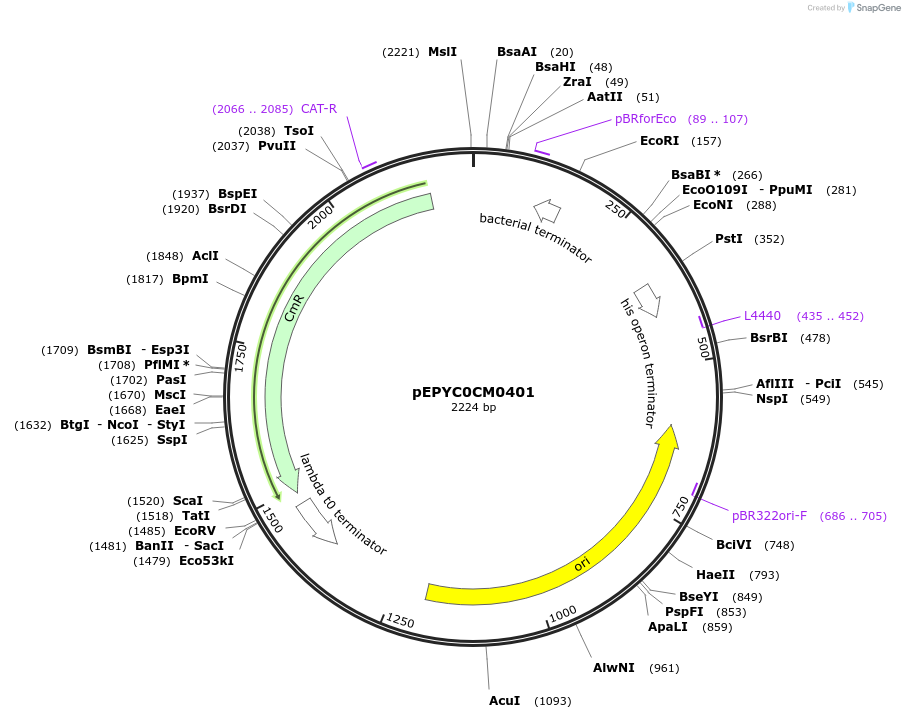
pEPYC0CM0401
Plasmid#154556PurposePhytobricks Level 0 partDepositorInsertMinSyn_060
UseSynthetic BiologyAvailable SinceOct. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
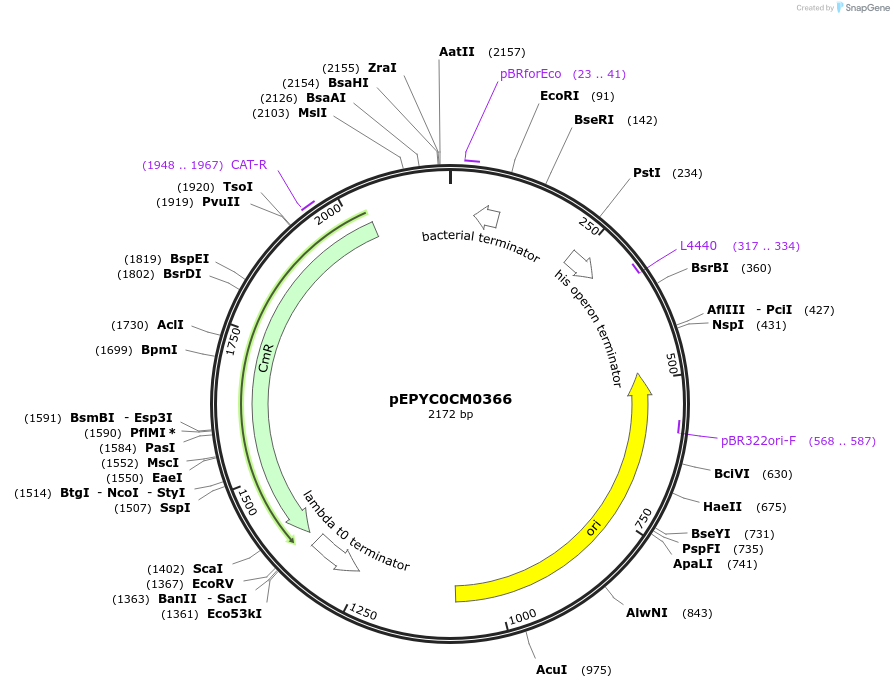
pEPYC0CM0366
Plasmid#154504PurposePhytobricks Level 0 partDepositorInsertMinSyn_001
UseSynthetic BiologyAvailable SinceOct. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
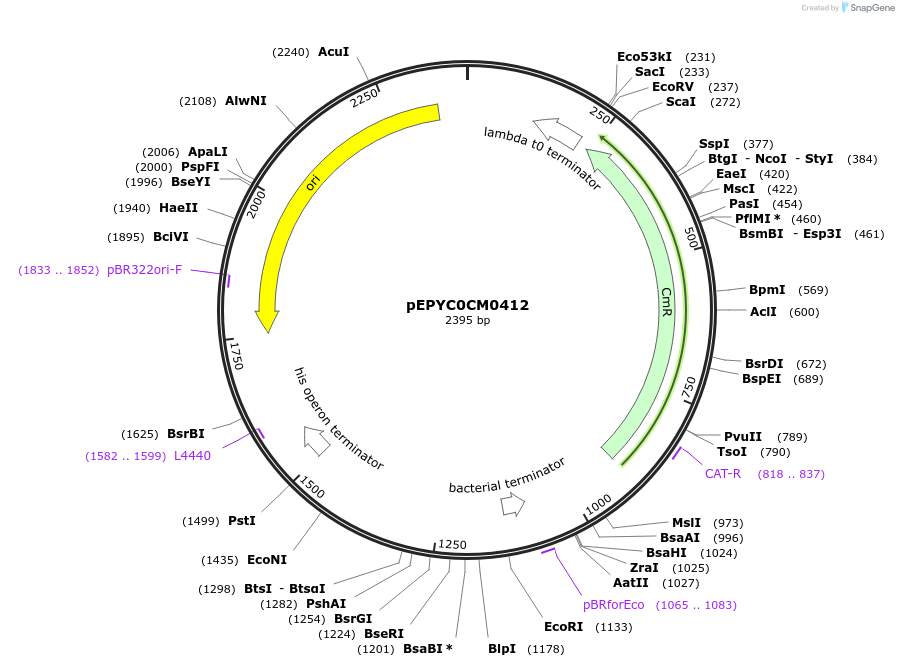
pEPYC0CM0412
Plasmid#154569PurposePhytobricks Level 0 partDepositorInsertMinSyn_106
UseSynthetic BiologyAvailable SinceOct. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
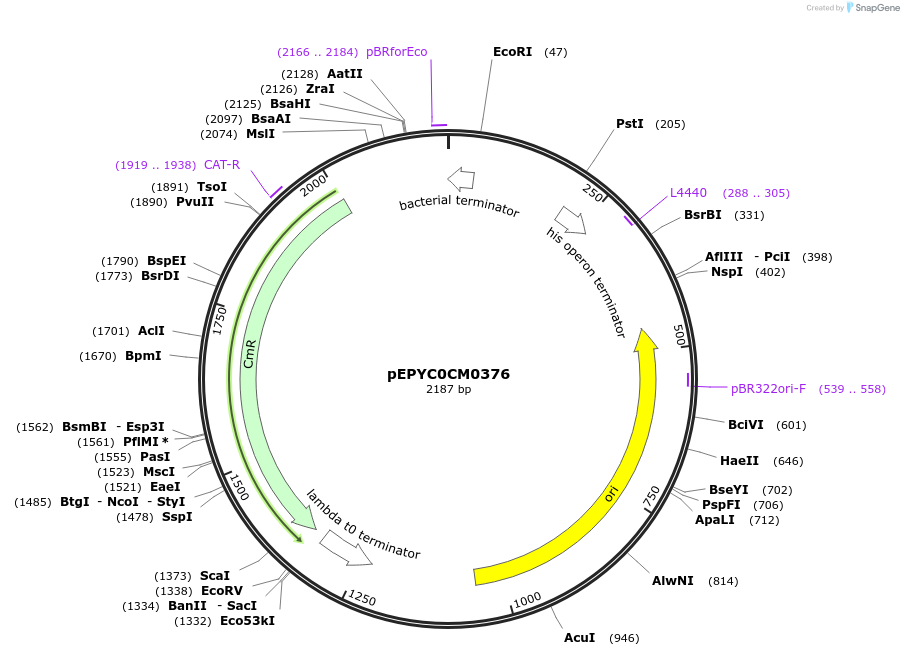
pEPYC0CM0376
Plasmid#154520PurposePhytobricks Level 0 partDepositorInsertMinSyn_017
UseSynthetic BiologyAvailable SinceOct. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
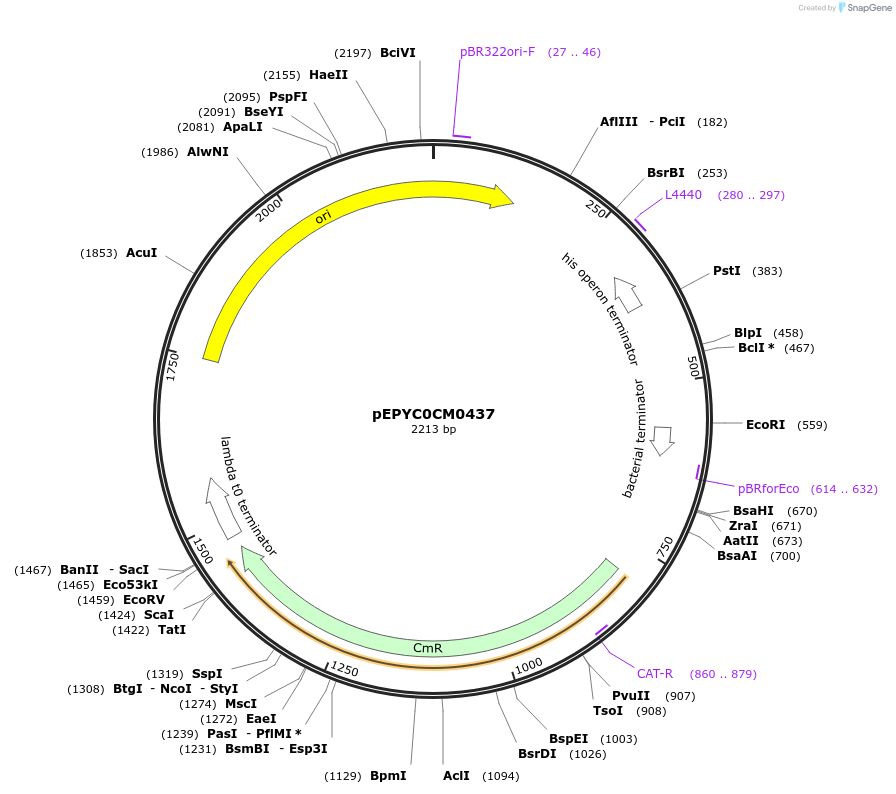
pEPYC0CM0437
Plasmid#154530PurposePhytobricks Level 0 partDepositorInsertMinSyn_028
UseSynthetic BiologyAvailable SinceOct. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
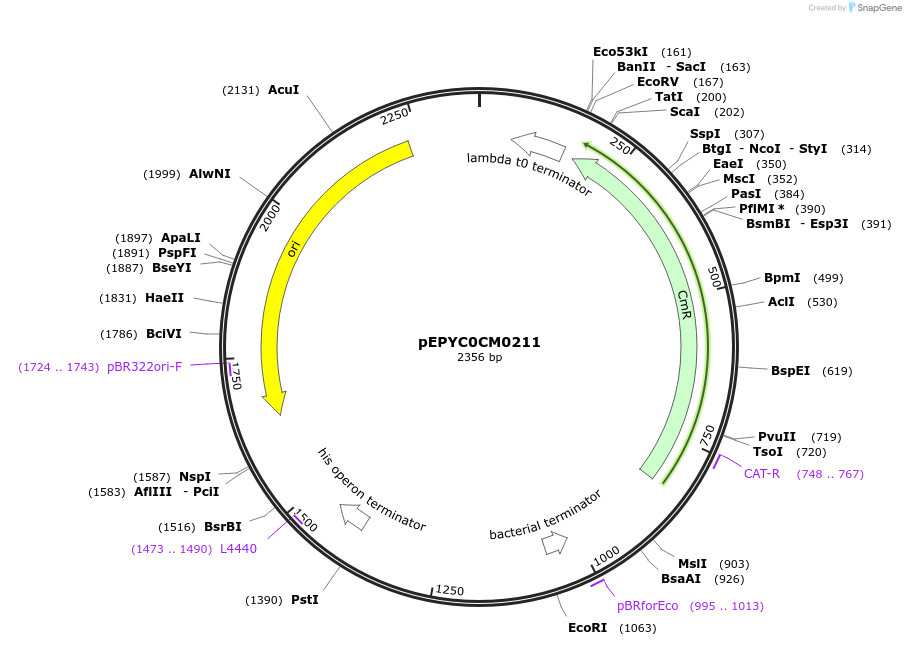
pEPYC0CM0211
Plasmid#154543PurposePhytobricks Level 0 partDepositorInsertMinSyn_041
UseSynthetic BiologyAvailable SinceOct. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
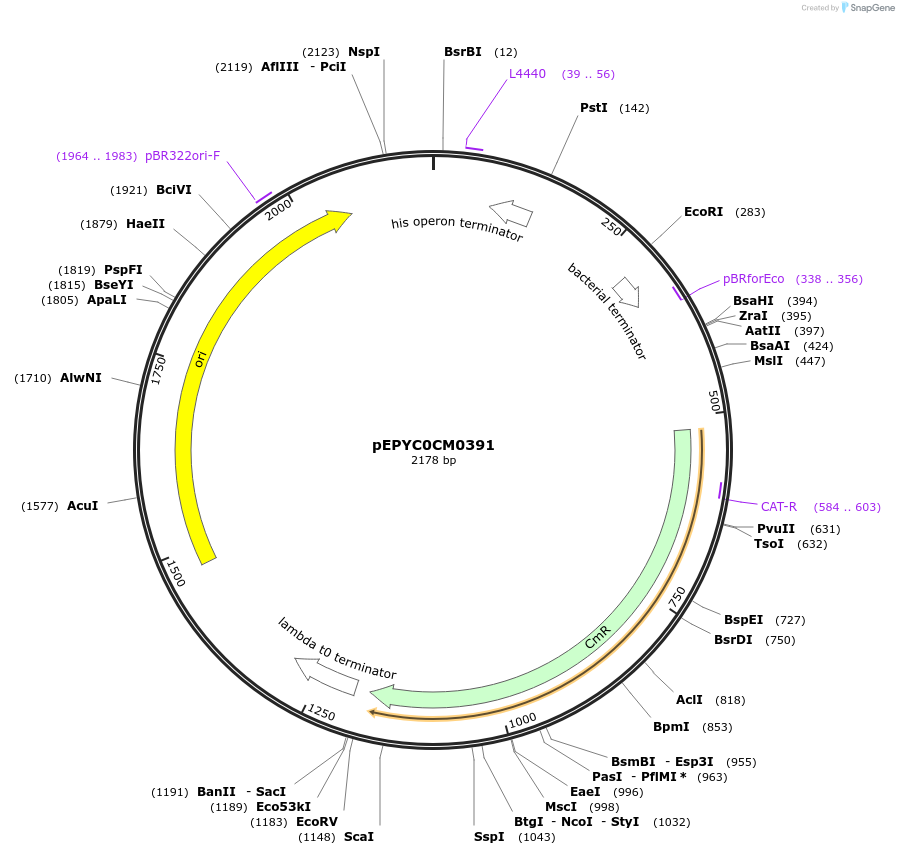
pEPYC0CM0391
Plasmid#154517PurposePhytobricks Level 0 partDepositorInsertMinSyn_014
UseSynthetic BiologyAvailable SinceOct. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
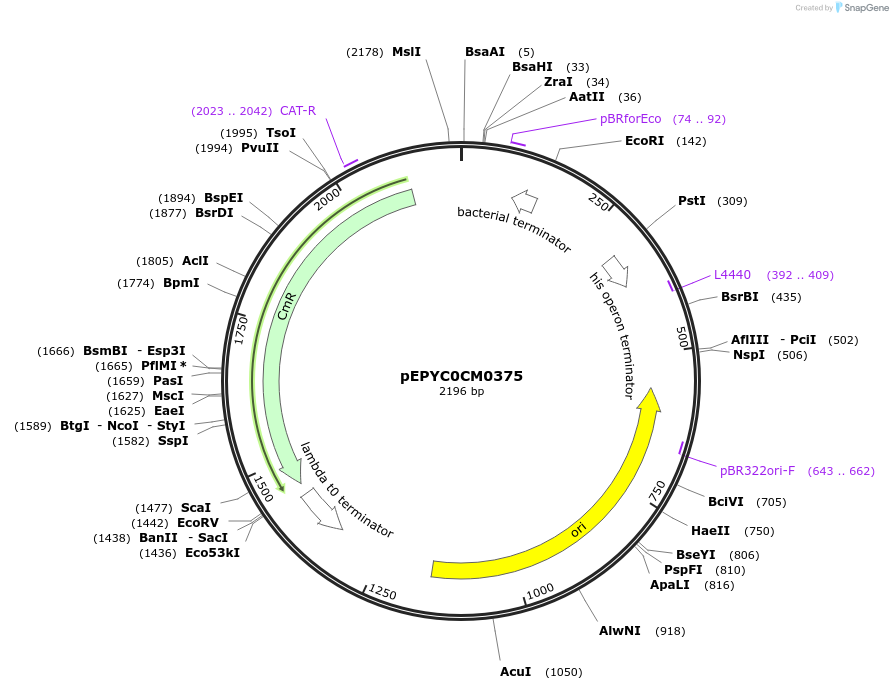
pEPYC0CM0375
Plasmid#154519PurposePhytobricks Level 0 partDepositorInsertMinSyn_016
UseSynthetic BiologyAvailable SinceOct. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
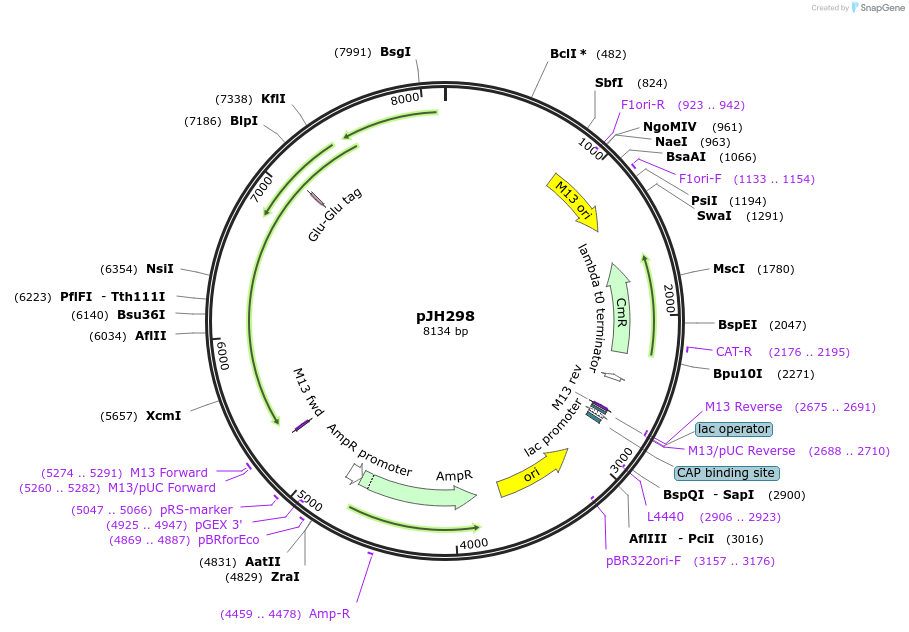
pJH298
Plasmid#115651Purposerecombination substrateDepositorInsertV(D)J recombination substrate
Available SinceSept. 24, 2018AvailabilityAcademic Institutions and Nonprofits only