We narrowed to 4,207 results for: Aes
-
Plasmid#213945PurposeAAV construct expressing mCherry-tagged channelrhodopsin-2 driven by SST interneuron-targeting enhancer. Alias: pAAV_WDS0004_ChR2_mCherryDepositorHas ServiceAAV PHP.eB and AAV1InsertChannelrhodopsin 2-mCherry
UseAAVAvailable SinceJuly 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
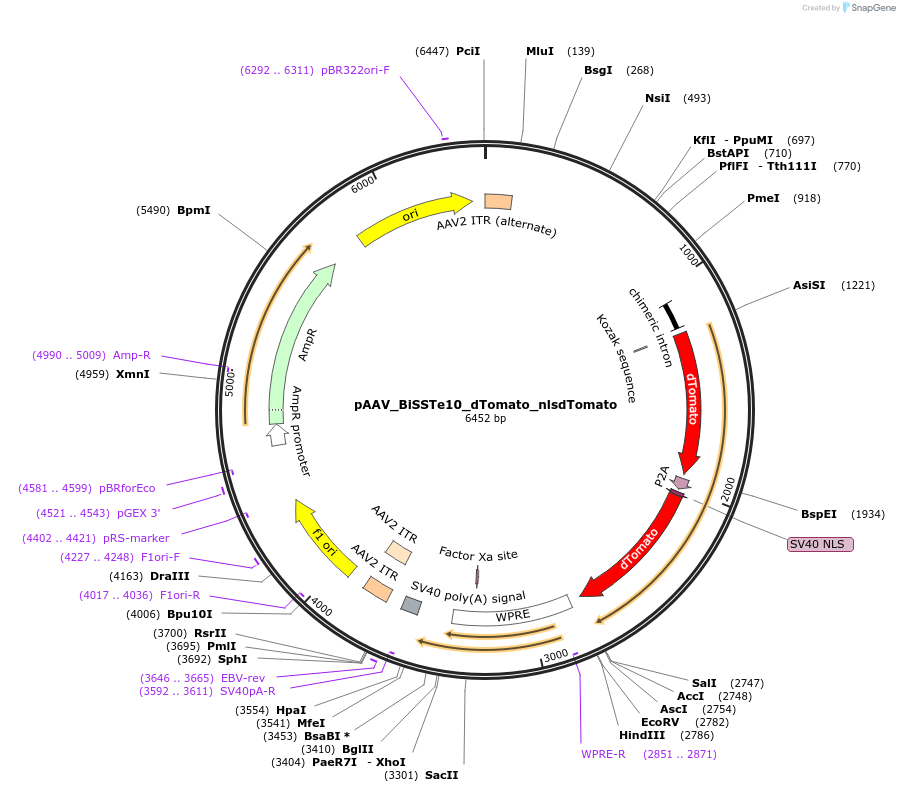
pAAV_BiSSTe10_dTomato_nlsdTomato
Plasmid#213814PurposeAAV construct expressing dTomato driven by SST interneuron-targeting enhancer. Alias: pAAV_S9E10_dTomato_nlsdTomatoDepositorHas ServiceAAV PHP.eB and AAV1InsertdTomato_P2A_nls.dTomato
UseAAVAvailable SinceJuly 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
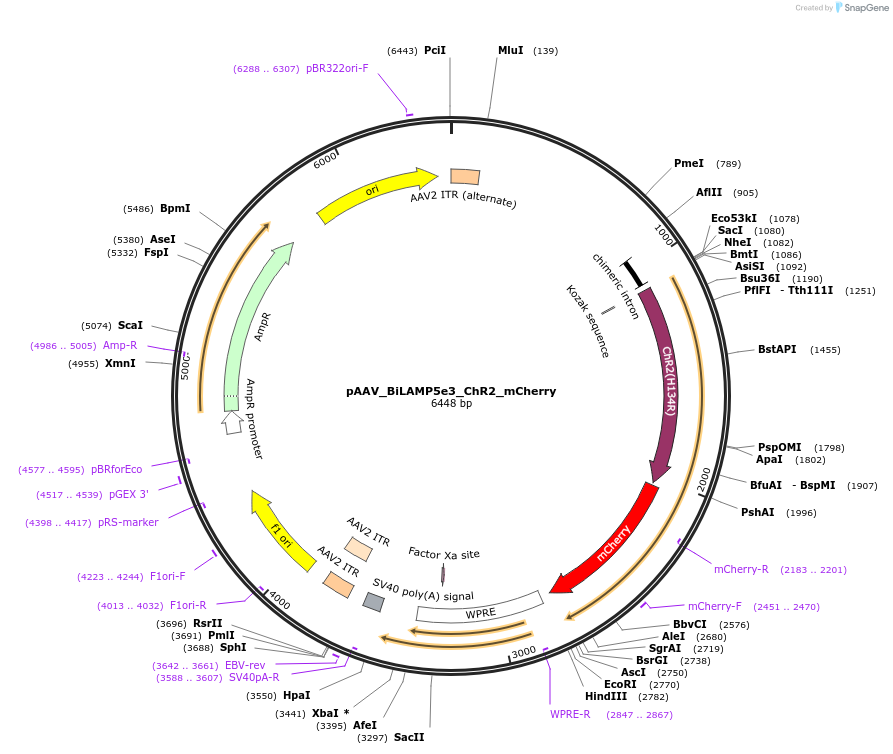
pAAV_BiLAMP5e3_ChR2_mCherry
Plasmid#213915PurposeAAV construct expressing mCherry-tagged channelrhodopsin-2 driven by Lamp5 interneuron-targeting enhancer. Alias: pAAV_WDL0003_ChR2_mCherryDepositorHas ServiceAAV PHP.eB and AAV1InsertChannelrhodopsin 2-mCherry
UseAAVAvailable SinceJuly 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
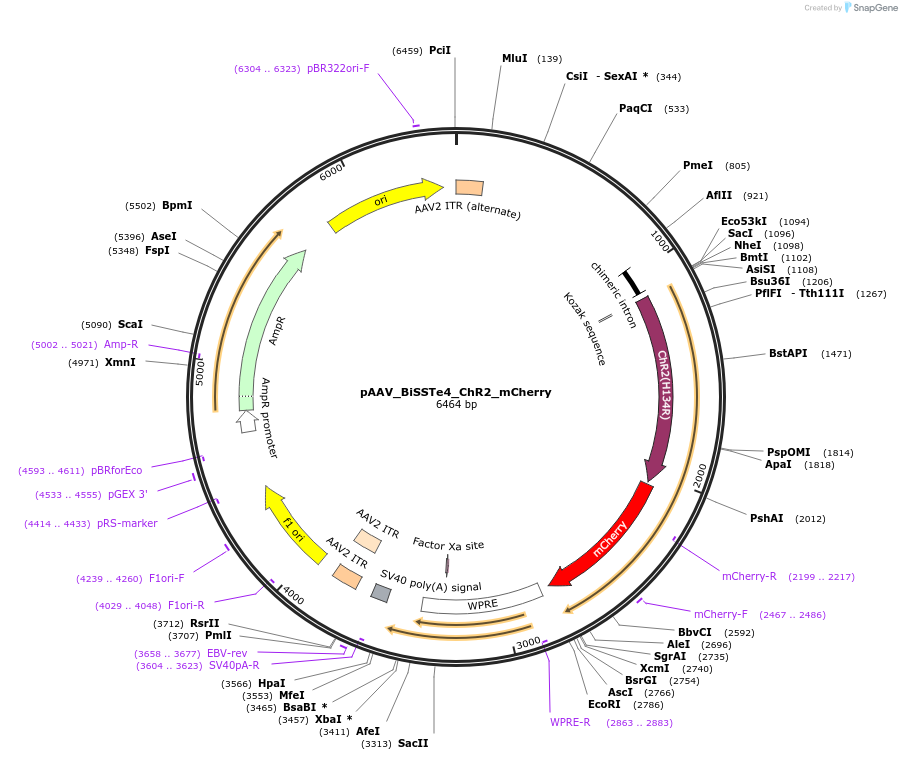
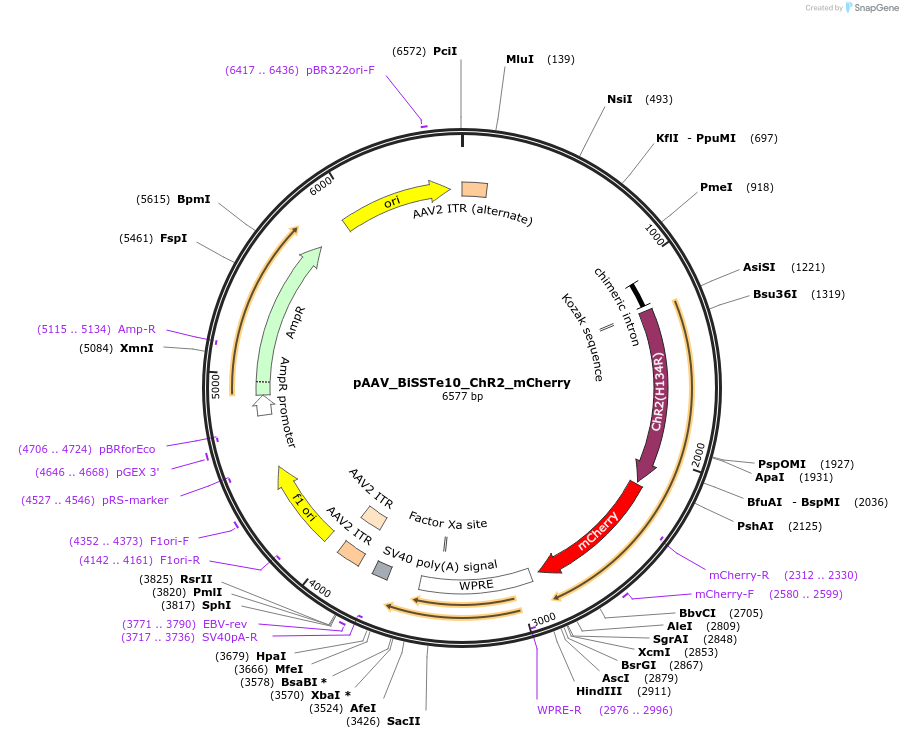
pAAV_BiSSTe10_ChR2_mCherry
Plasmid#213815PurposeAAV construct expressing mCherry-tagged channelrhodopsin-2 driven by SST interneuron-targeting enhancer. Alias: pAAV_S9E10_ChR2_mCherryDepositorHas ServiceAAV PHP.eB and AAV1InsertChannelrhodopsin 2-mCherry
UseAAVAvailable SinceJuly 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
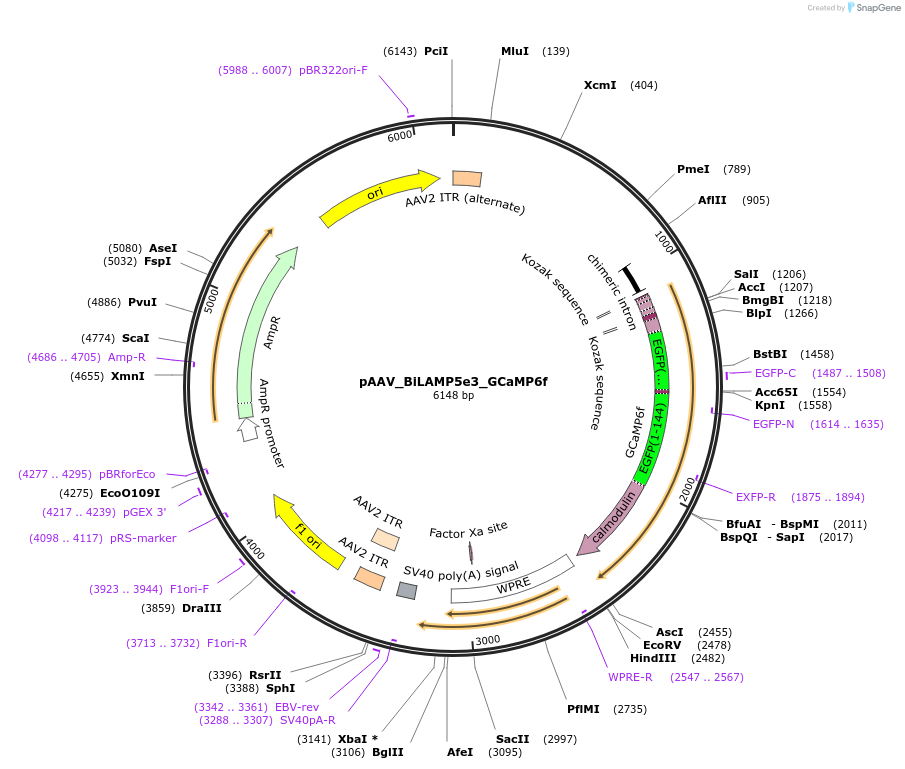
pAAV_BiLAMP5e3_GCaMP6f
Plasmid#213917PurposeAAV construct expressing GCaMP6f driven by Lamp5 interneuron-targeting enhancer. Alias: pAAV_WDL0003_GCaMP6fDepositorHas ServiceAAV1InsertGCaMP6f
UseAAVAvailable SinceJuly 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
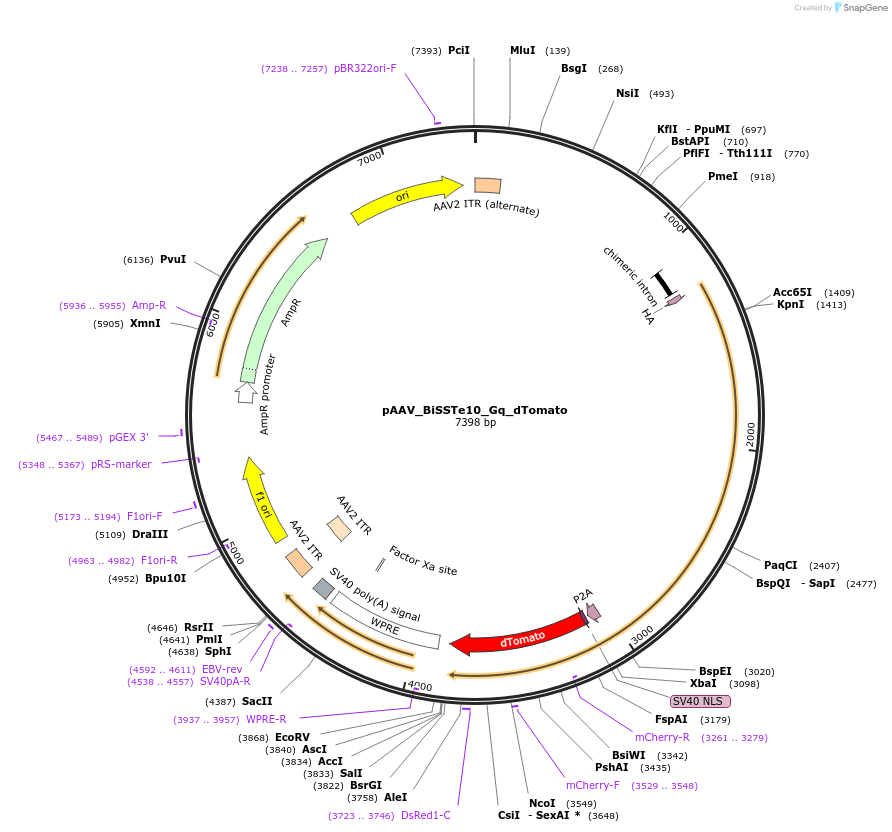
pAAV_BiSSTe10_Gq_dTomato
Plasmid#213817PurposeAAV construct expressing HA-hM3Dq-dTomato driven by SST interneuron-targeting enhancer. Alias: pAAV_S9E10_Gq_dTomatoDepositorInsertHA-GqDREADD-dTomato
UseAAVAvailable SinceJuly 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
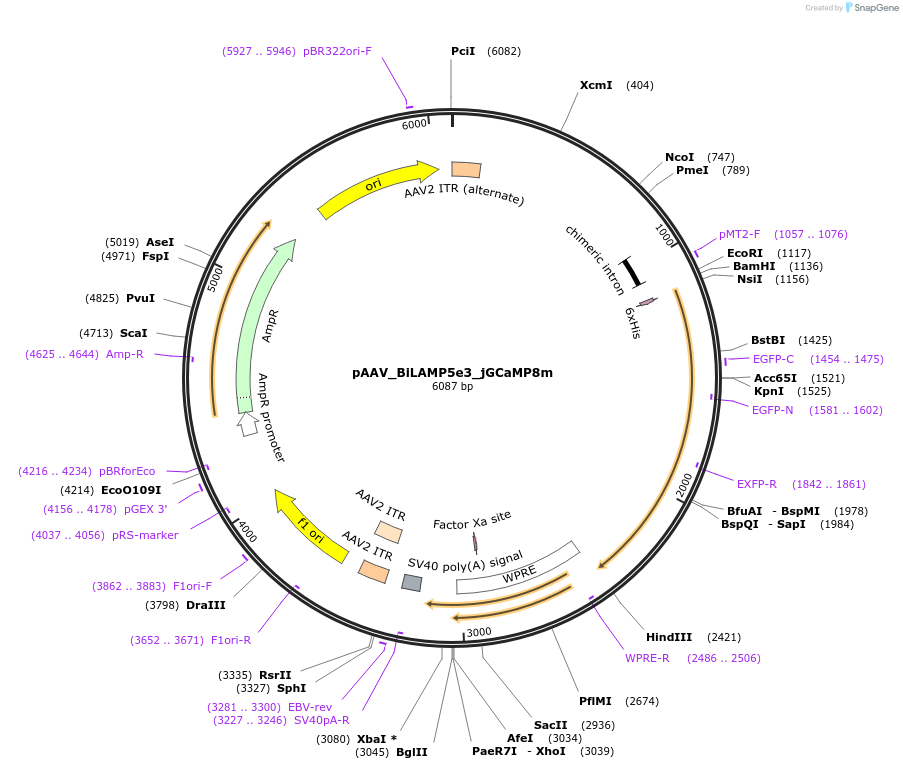
pAAV_BiLAMP5e3_jGCaMP8m
Plasmid#213918PurposeAAV construct expressing jGCaMP8m driven by Lamp5 interneuron-targeting enhancer. Alias: pAAV_WDL0003_jGCaMP8mDepositorHas ServiceAAV1InsertjGCaMP8m
UseAAVAvailable SinceJuly 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
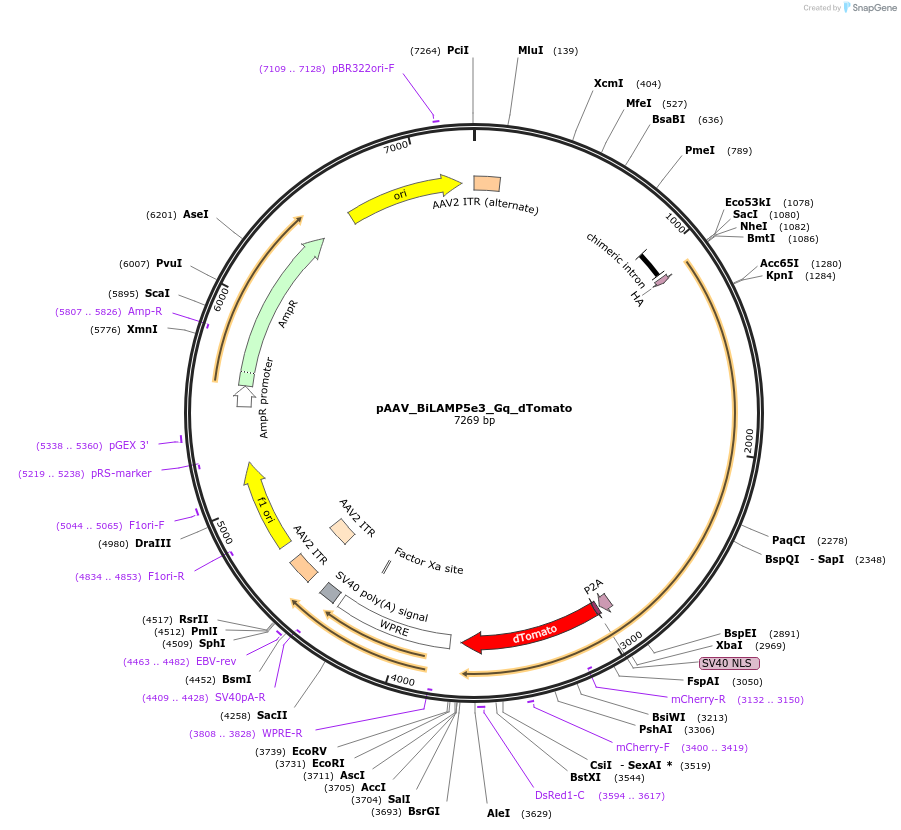
pAAV_BiLAMP5e3_Gq_dTomato
Plasmid#213916PurposeAAV construct expressing HA-hM3Dq-dTomato driven by Lamp5 interneuron-targeting enhancer. Alias: pAAV_WDL0003_Gq_dTomatoDepositorInsertHA-GqDREADD-dTomato
UseAAVAvailable SinceJuly 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
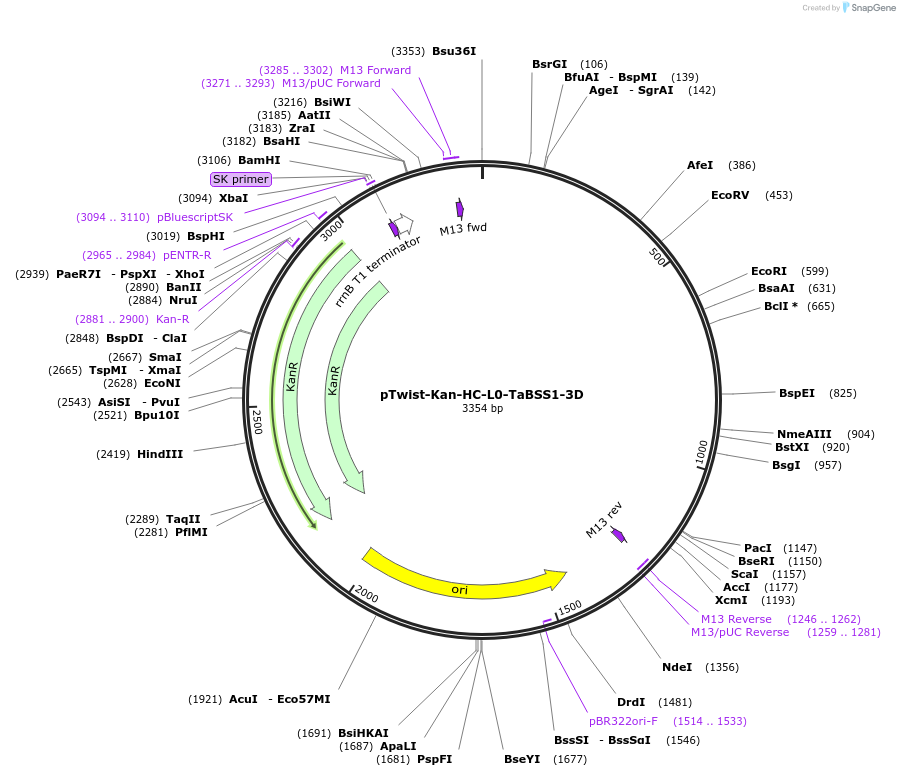
pTwist-Kan-HC-L0-TaBSS1-3D
Plasmid#242008PurposeL0 3U* Golden Gate part of 3' UTR and downstream regulatory regions of Triticum aestivum MANY-NODED DWARF 1 (MND1) B-subgenome homoeolog (TraesCS7B02G413900)DepositorInsertTaBSS1-3D
UseSynthetic BiologyExpressionPlantAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
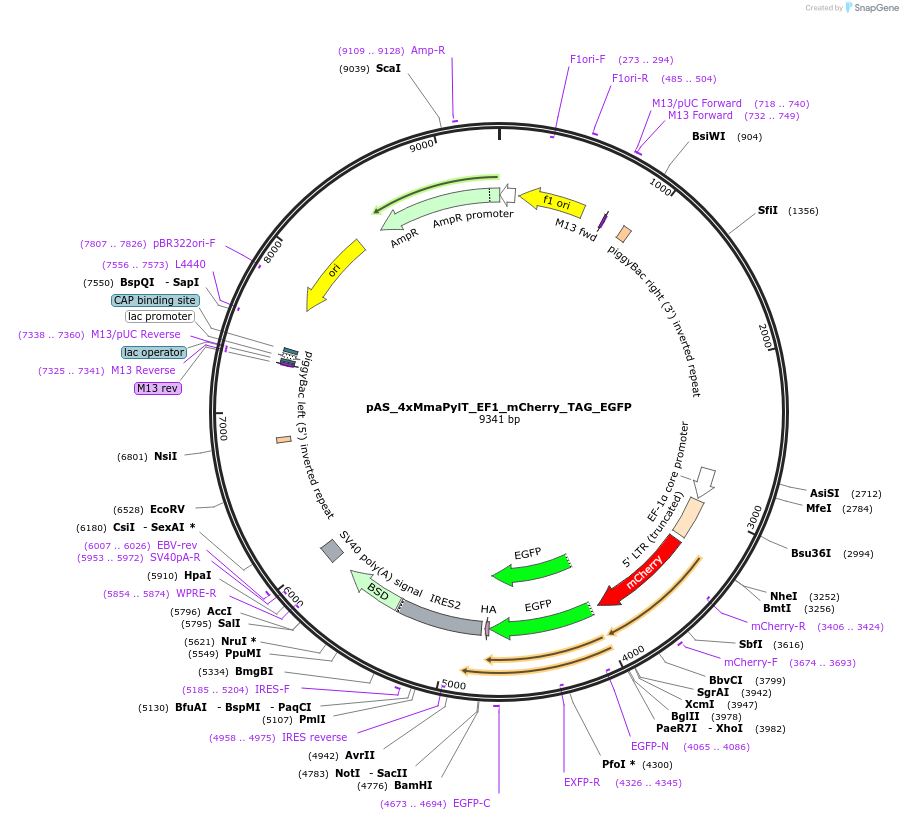
pAS_4xMmaPylT_EF1_mCherry_TAG_EGFP
Plasmid#174893Purposeamber suppression reporter mCherry-TAG-EGFP expression, with MmaPylT amber suppressor tRNA cassetteDepositorInsertmCherry and EGFP
ExpressionMammalianPromoterEF1Available SinceNov. 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
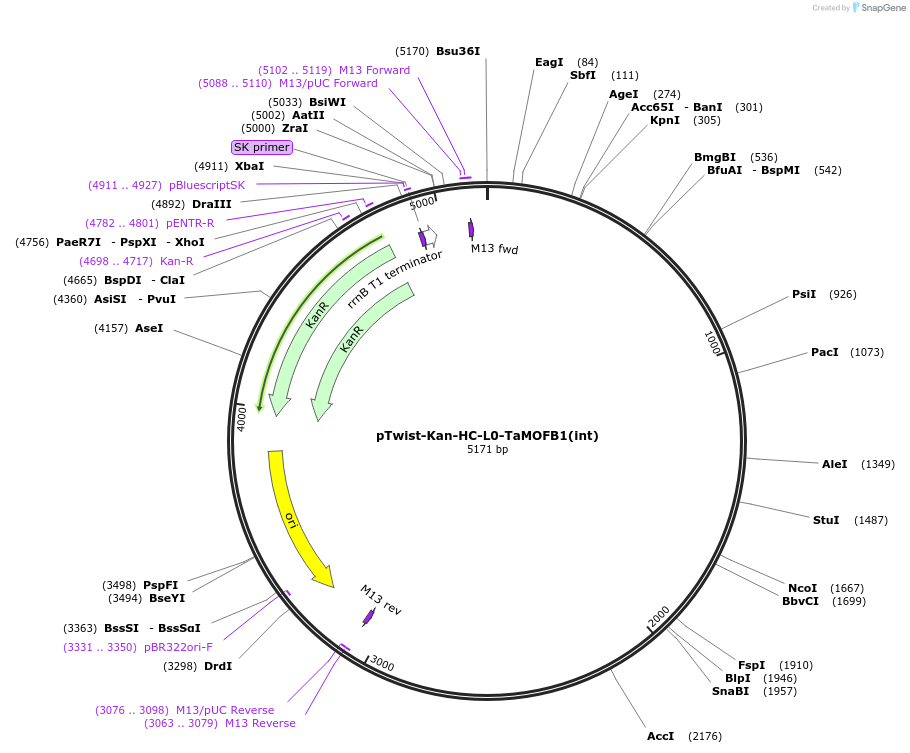
pTwist-Kan-HC-L0-TaMOFB1(int)
Plasmid#242005PurposeL0 CDS1 Golden Gate part of the pre-mRNA (excluding UTRs) of Triticum aestivum MORE FLORET 1 (MOF-1) / MULTI-FLORET SPIKELET 2 (MFS-2) B-subgenome homoeolog (TraesCS2B02G420900)DepositorInsertTaMOF-B1_with_introns
UseSynthetic BiologyExpressionPlantAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
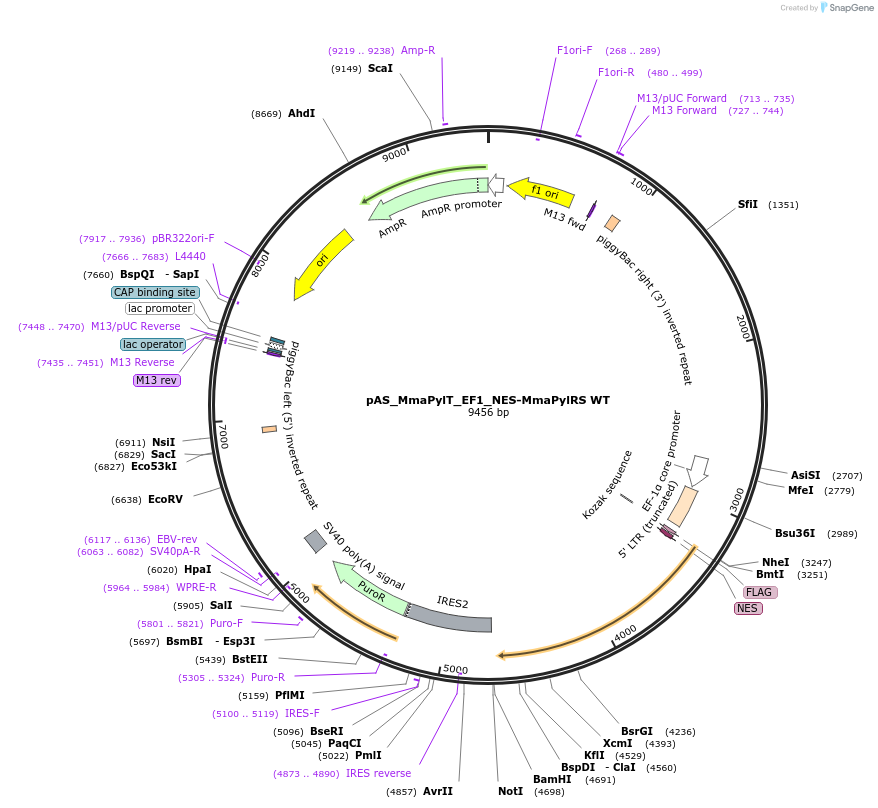
pAS_MmaPylT_EF1_NES-MmaPylRS WT
Plasmid#174901Purposeexpression of Mma PylRS with nuclear export sequence for amber suppression in mammalian cells; transient or piggy bac mediated integrationDepositorAvailable SinceNov. 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
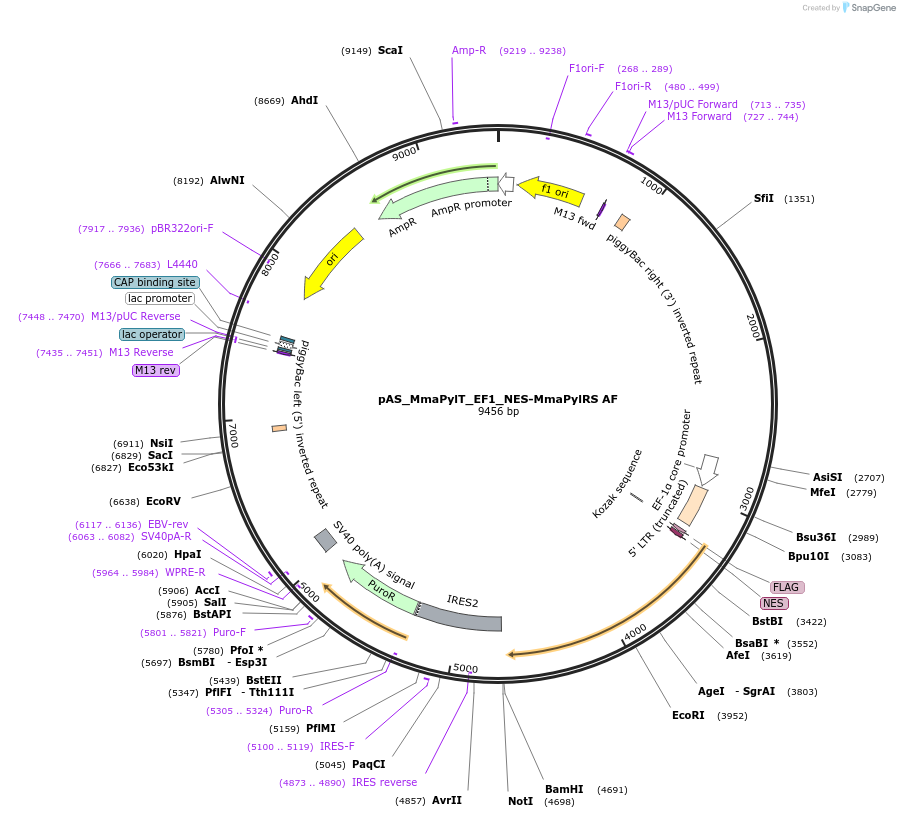
pAS_MmaPylT_EF1_NES-MmaPylRS AF
Plasmid#174902Purposeexpression of Mma PylRS AF with nuclear export sequence for amber suppression in mammalian cells; transient or piggy bac mediated integrationDepositorInsertMma PylRS (pylS Methanosarcina mazei)
TagsFLAG and NESExpressionMammalianMutationY306A, Y384FPromoterEF1Available SinceNov. 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
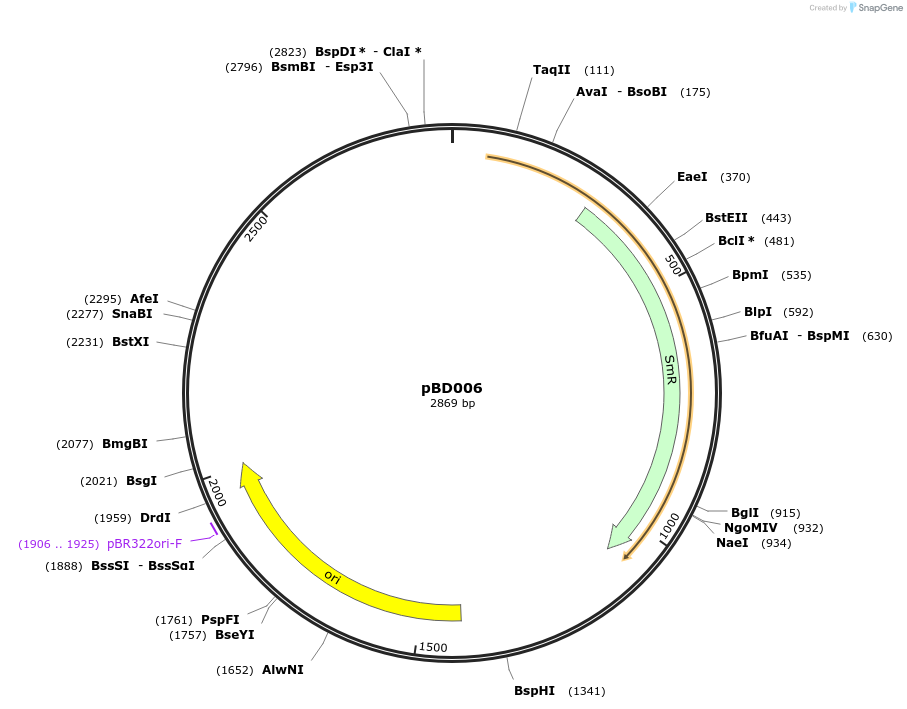
pBD006
Plasmid#201306PurposeD16: Promoter of TraesCS5D02G320700 as Golden Gate compatible MoClo PROM partDepositorInsertD16: Promoter of TraesCS5D02G320700
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceJan. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
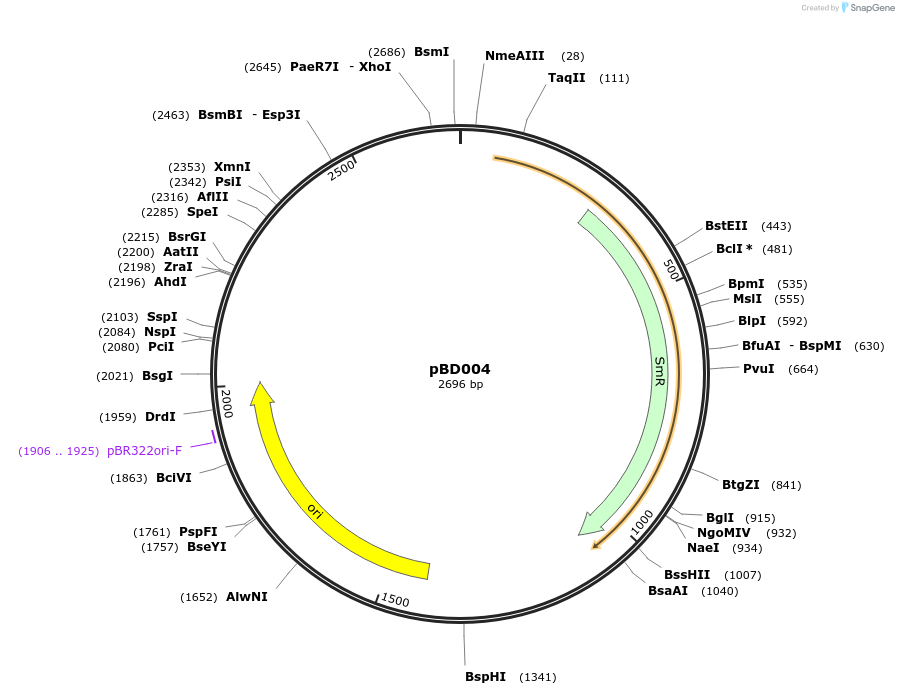
pBD004
Plasmid#201304PurposeD14: Promoter of TraesCS3D02G368800 as Golden Gate compatible MoClo PROM partDepositorInsertD14: Promoter of TraesCS3D02G368800
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
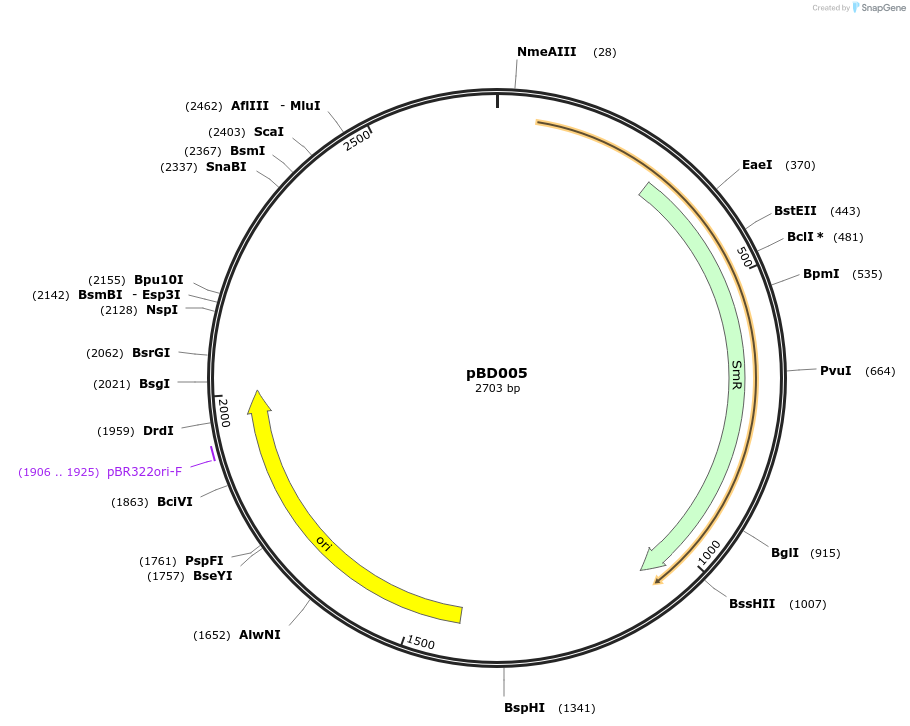
pBD005
Plasmid#201305PurposeD15: Promoter of TraesCS3D02G517500 as Golden Gate compatible MoClo PROM partDepositorInsertD15: Promoter of TraesCS3D02G517500
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
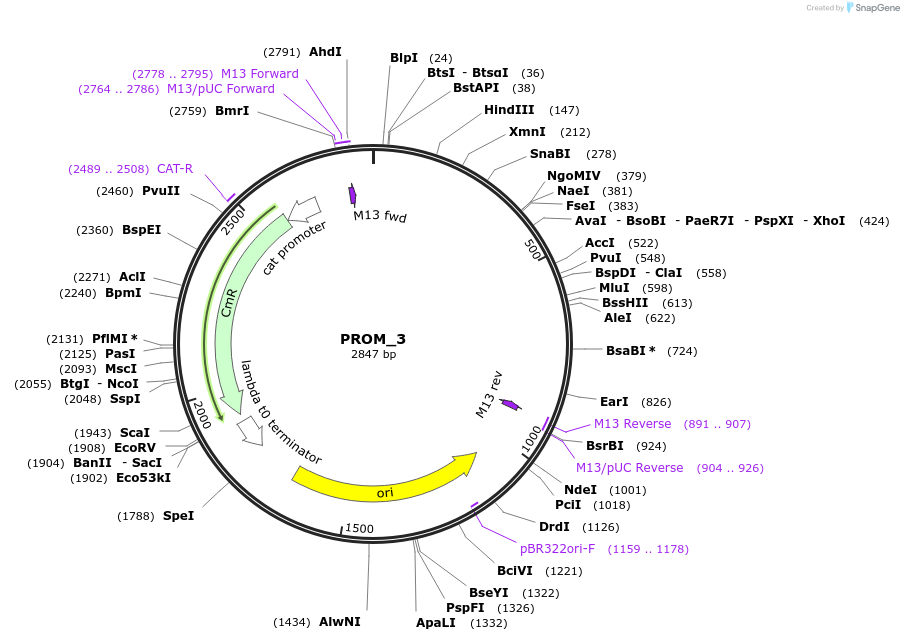
PROM_3
Plasmid#201296PurposeD3: Promoter of TraesCS3A02G510700 as Golden Gate compatible MoClo PROM partDepositorInsertD3: Promoter of TraesCS3A02G510700
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
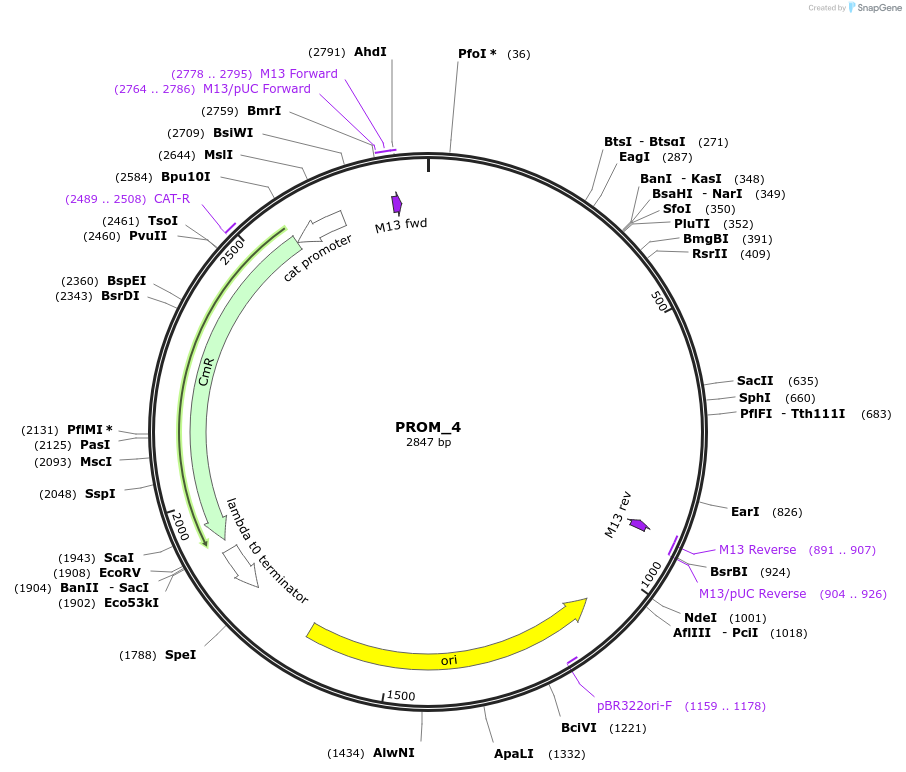
PROM_4
Plasmid#201297PurposeD4: Promoter of TraesCS3B02G552700 as Golden Gate compatible MoClo PROM partDepositorInsertD4: Promoter of TraesCS3B02G552700
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
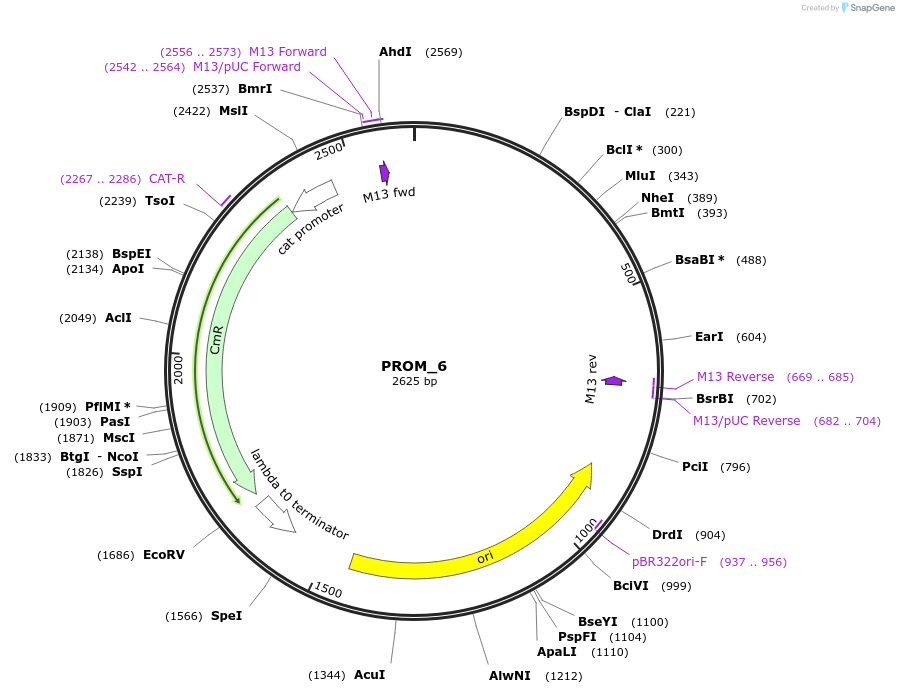
PROM_6
Plasmid#201299PurposeD6: Promoter of TraesCS3B02G578800 as Golden Gate compatible MoClo PROM partDepositorInsertD6: Promoter of TraesCS3B02G578800
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
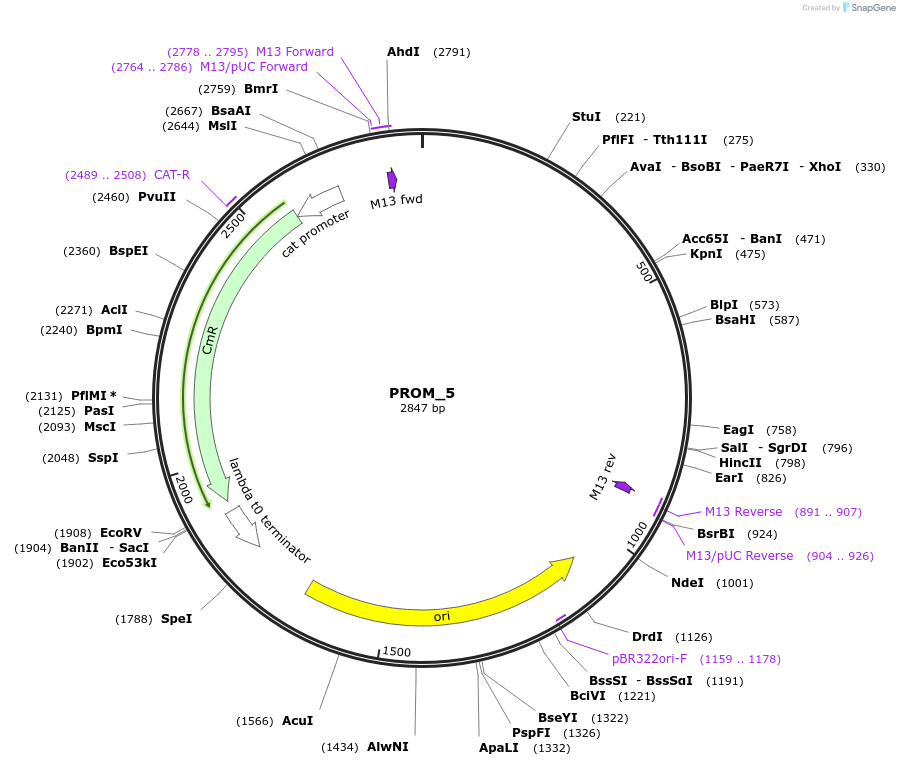
PROM_5
Plasmid#201298PurposeD5: Promoter of TraesCS3B02G553900 as Golden Gate compatible MoClo PROM partDepositorInsertD5: Promoter of TraesCS3B02G553900
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only