CHyMErA Cas12a Nuclease Optimization, Large-Scale Exon Deletion, and scCHyMErA-Seq Exon Deletion hgRNA Libraries
(Pooled Libraries #209734, #209735, #209736, #237473)
-
Purpose
Cas Hybrid for Multiplexed Editing and screening Applications (CHyMErA) lentiviral libraries express hybrid Cas9-Cas12a guide RNAs under a single U6 promoter.
The CHyMErA Cas12a Nuclease Optimization hybrid guide (hgRNA) libraries allow the user to assess and compare the efficacy of different LbCas12a (#209734) or AsCas12a nucleases (#209735) in the context of the Cas9-Cas12a CHyMErA system (Gonatopoulos-Pournatzis et al., 2020).
The CHyMErA Large-Scale Exon Deletion hgRNA library (#209736) screens for frame-preserving exons that affect cell fitness in human cells, using the CHyMErA screening platform (Gonatopoulos-Pournatzis et al., 2020) with SpCas9 and AsCas12a nucleases.
The scCHyMErA-Seq Exon Deletion hgRNA library (#237473) enables high-throughput identification of frame-preserving exons that influence transcriptional phenotypes, building on the CHyMErA system (Nature Biotechnology, 2020; PMID: 32249828). It uses SpCas9 and (As)opCas12a nucleases for combinatorial exon deletion and is compatible with 10x Genomics single-cell transcriptomics. By integrating exon deletions with single-cell RNA-Seq readouts, this tool enables studying the impact of the targeted exons on transcriptional phenotypes.
-
Vector Backbone
#209734 and #209735: pLCHKOv2 - does not express Cas9 or Cas12a
#209736: pLCHKOv3 (Plasmid #209046) - does not express Cas9 or Cas12a
#237473: pLCHKOv4 (Plasmid #237451) - does not express Cas9 or Cas12a
-
Depositing Labs
-
Publication
#209734, #209735, #209736:
Xiao et al. Mol Cell. 2024 June 24. doi: 10.1016/j.molcel.2024.05.024. (How to cite )#237473:
Gonatopoulos-Pournatzis et al. (Unpublished) (How to cite )
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |||
|---|---|---|---|---|---|---|---|
| Pooled Library | 209734 | CHyMErA LbCas12a Nuclease Optimization hgRNA library† | 1 | $473 | Add to Cart | ||
| Pooled Library | 209735 | CHyMErA AsCas12a Nuclease Optimization hgRNA library† | 1 | $473 | Add to Cart | ||
| Pooled Library | 209736 | CHyMErA Large-Scale Exon Deletion hgRNA library† | 1 | $935 | Add to Cart | ||
| Pooled Library | 237473 | scCHyMErA-Seq Exon Deletion library 1†† | 1 | $418 | Add to Cart | ||
Library Details
| Pooled Library | Genes Targeted | gRNAs / element | hgRNAs |
|---|---|---|---|
| 209734 | 844 | 1–10 | 18,000 |
| 209735 | 844 | 1–10 | 18,000 |
| 209736 | 12,126 exons 2,095 genes |
6–18 | 300,000 |
| 237473 | 224 exons 161 genes |
3 per exon 2 per gene |
1,066 |
-
SpeciesHuman
-
Controls
#209734–209736: 1,503 dual intergenic and 100 non-targeting hgRNAs in each library
#237473: 40 intergenic and 40 non-targeting hgRNAs -
Lentiviral Generation3rd
Library Shipping
This library is delivered as suspended DNA in a microcentrifuge tube on blue ice. The tube's contents will not necessarily be frozen. For best results, minimize freeze-thaws.
- For #209734, #209735, and #209736
- Volume: ∼20 µL
- Concentration: 50 ng/µL
- For #237473
- Volume: ∼15 µL
- Concentration: 50 ng/µL
Resource Information
-
Protocols
-
Depositor Data
- #209734 Distribution of NGS Reads (PDF, 149 KB)
- #209735 Distribution of NGS Reads (PDF, 126 KB)
- #209734 and #209735 hgRNA sequences (XLSX, 3.1 MB)
- #209736 Distribution of NGS Reads (PDF, 124 KB)
- #209736 hgRNA sequences (XLSB, 25.2 MB)
- #237473 Distribution of NGS Reads (PDF, 125 KB)
- #237473 hgRNA sequences (XLSX, 213 KB)
-
Scripts
For #209734, #209735, and #209736 -
Terms and Licenses
Academic/Nonprofit Terms
Depositor Comments
CHyMErA LbCas12a and AsCas12a Nuclease Optimization hgRNA Libraries
The Cas12a Nuclease Optimization hgRNA libraries can be used to assess dropout and enrichment of guides targeting reference core essential and nonessential genes via single and combinatorial genome editing. Each nuclease optimization hgRNA library consists of 18,000 hgRNAs targeting 482 core essential and 362 nonessential genes through gene knockout (9,129 hgRNAs) or deletion of frame-disruptive exons (3,236 hgRNAs). In addition, the libraries contain controls including intronic-intergenic (4,032 hgRNAs), dual intergenic (1,503 hgRNAs) and non-coding (100 hgRNAs) targeting hgRNAs.
CHyMErA Large-Scale Exon Deletion hgRNA Library
The Large-Scale Exon Deletion hgRNA library aims to delete all targetable frame-preserving exons in DepMap (Link opens in a new window) common essential genes (5,333 exons; 85,188 hgRNAs) along with 2,489 frame-preserving exons in additional genes (41,044 hgRNAs). This library also targets 4,304 frame-disruptive exons (i.e., exons whose deletion results in gene inactivation) in common essential and additional genes (42,560 hgRNAs) to evaluate the gene phenotype. The library contains all hgRNAs included in the CHyMErA Cas12a Nuclease Optimization hgRNA libraries to determine the efficacy of Cas9 and Cas12a for gene knockout and exon deletion.

Figure 1: CHyMErA system overview. Cells express SpCas9 and either LbCas12a or AsCas12a and an hgRNA fusion of Cas9 and Cas12a gRNAs. Cas12a RNA processing activity generates individual Cas9 and Cas12a gRNAs which bind to their respective nucleases and effect combinatorial targeting.
scCHyMErA-Seq Exon Deletion Library 1
The scCHyMErA-Seq Exon Deletion Library 1 is a scalable CRISPR-based exon deletion screening platform integrated with 10x Genomics single-cell transcriptomic readouts. This tool enables efficient exon deletion while simultaneously capturing Cas9/Cas12a guides and polyadenylated transcripts at single-cell resolution. Importantly, gene expression profiles generated using scCHyMErA-Seq accurately recapitulate findings from traditional, labor-intensive orthogonal methods, while offering enhanced scalability and efficiency. The library includes 1,066 hgRNAs targeting 224 exons in 161 genes, with three independent hgRNAs for each exon deletion. It also provides gene knockout hgRNAs (two per gene), with 40 intergenic and 40 non-targeting hgRNAs as controls. Overall, scCHyMErA-Seq represents a robust and versatile platform for systematically unraveling the functional impact of alternative splicing by directly linking specific splicing variants to transcriptional phenotypes.
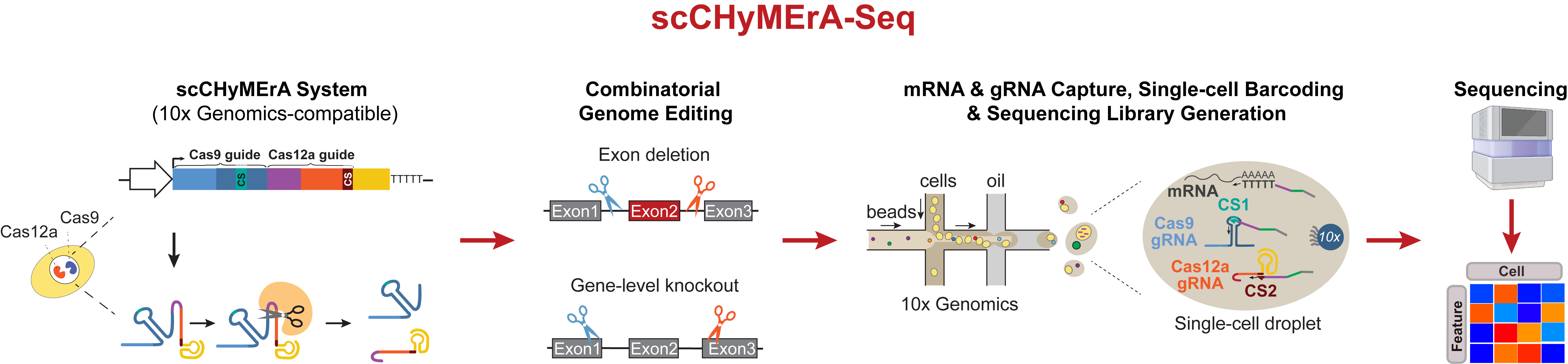
Figure 2: Workflow of the scCHyMErA-seq system. Exon deletion screening is coupled with 10x Genomics single-cell transcriptomics, enabling simultaneous capture of gRNAs mediating exon deletions together with mRNA transcripts at the single-cell level. Integrative analysis links specific exon deletions to transcriptional phenotypes. Capture sequence 1 (CS1) and capture sequence 2 (CS2) within the single-cell droplet allow detection of Cas9 and Cas12a gRNAs, respectively, corresponding to cut site 1 and cut site 2 in the genome.
These pooled libraries were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
The CHyMErA LbCas12a Nuclease Optimization hgRNA Library was a gift from Thomas Gonatopoulos-Pournatzis (Addgene 209734 ; http://n2t.net/addgene:209734 ; RRID:Addgene_209734).
The CHyMErA AsCas12a Nuclease Optimization hgRNA Library was a gift from Thomas Gonatopoulos-Pournatzis (Addgene 209735 ; http://n2t.net/addgene:209735 ; RRID:Addgene_209735)
The CHyMErA Large-Scale Exon Deletion hgRNA Library was a gift from Thomas Gonatopoulos-Pournatzis (Addgene 209736 ; http://n2t.net/addgene:209736 ; RRID:Addgene_209736)
The scCHyMErA-Seq Exon Deletion Library 1 was a gift from Thomas Gonatopoulos-Pournatzis (Addgene 237473 ; http://n2t.net/addgene:237473 ; RRID:Addgene_237473) -
For your References section:
For #209734–209736:
Genome-scale exon perturbation screens uncover exons critical for cell fitness. Xiao M-S, Damodaran AP, Kumari B, Dickson E, Xing K, On TA, Parab N, King HE, Perez AR, Guiblet WM, Duncan G, Che A, Chari R, Andresson T, Vidigal JA, Weatheritt RJ, Aregger M, Gonatopoulos-Pournatzis T. Mol Cell. 2024 Jul 11;84(13):2553-2572.e19. doi: 10.1016/j.molcel.2024.05.024. Epub 2024 Jun 24. PubMed 38917794(Link opens in a new window)
For #237473:
A Scalable Exon Deletion Platform Coupled To Single-Cell Transcriptomics Reveals Exons Shaping Gene Expression Dynamics. Gonatopoulos-Pournatzis T et al. Unpublished.


