-
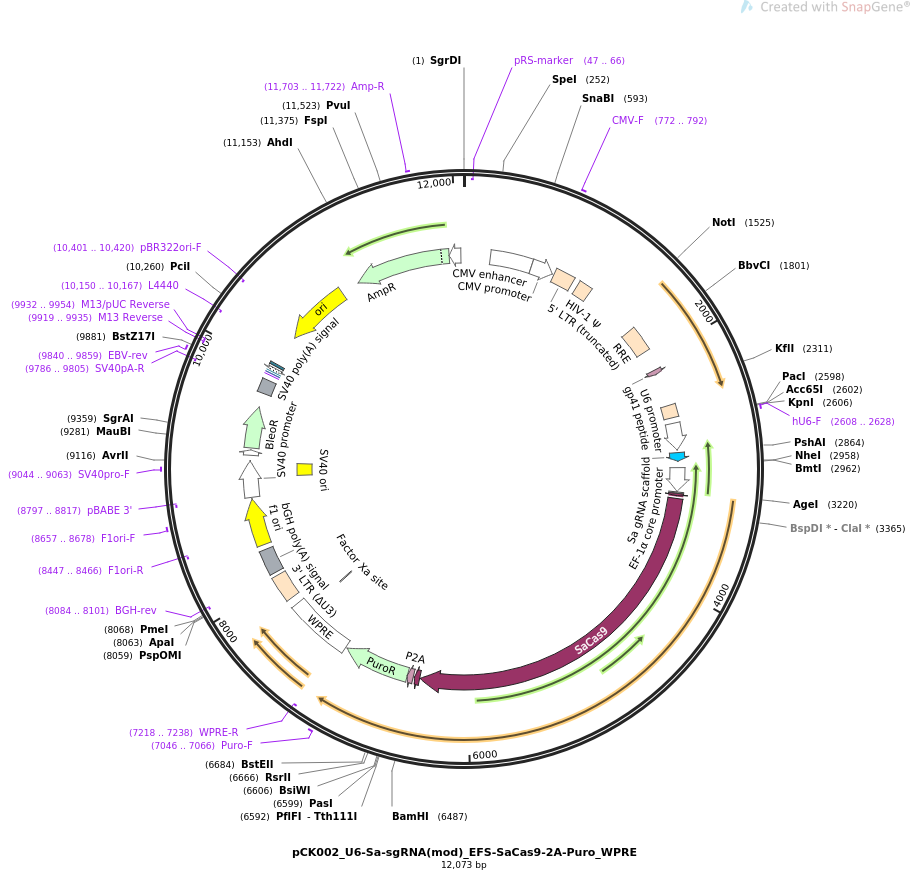
PurposeLentiviral vector encoding modified SaCas9 system backbone bearing BsmBI site for new guide RNAs and puromycin selection marker.
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 85452 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
This material is available to academics and nonprofits only.
Backbone
-
Vector backbonepLKO
- Backbone size w/o insert (bp) 6927
- Total vector size (bp) 12075
-
Vector typeMammalian Expression, Lentiviral, CRISPR
-
Selectable markersPuromycin
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)NEB Stable
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameU6-modified-sgRNA-Backbone-EFS-hSaCas9-2xNLS-2A-Puro
-
Alt nameSauCas9, sgRNA scaffold
-
SpeciesSynthetic; S aureus
-
Insert Size (bp)5148
-
Tags
/ Fusion Proteins
- NLS (N terminal on insert)
- NLS (C terminal on insert)
Resource Information
-
Article Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Any issue or question regarding this vector can be also directed to Le Cong (Regev lab) at [email protected].
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pCK002_U6-Sa-sgRNA(mod)_EFS-SaCas9-2A-Puro_WPRE was a gift from Aviv Regev (Addgene plasmid # 85452 ; http://n2t.net/addgene:85452 ; RRID:Addgene_85452) -
For your References section:
A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, Nyman J, Sakuishi K, Kurtulus S, Gennert D, Xia J, Kwon JY, Nevin J, Herbst RH, Yanai I, Rozenblatt-Rosen O, Kuchroo VK, Regev A, Anderson AC. Cell. 2016 Sep 8;166(6):1500-1511.e9. doi: 10.1016/j.cell.2016.08.052. 10.1016/j.cell.2016.08.052 PubMed 27610572