Guide to Plasmid Pooled Libraries
Pooled libraries represent a powerful tool for forward genetic screening or identifying previously unknown genes that contribute to a phenotype. Pooled libraries consist of a single mixture of many different plasmids. Plasmids within a given library have the same backbone, but contain unique inserts. Some libraries target or express genes representing the majority of a genome, while others are restricted to certain gene sets. Pooled libraries can vary greatly in size, from less than a hundred plasmids to millions. A well-designed screen can help you begin to understand what genes are important to a certain phenotype, and allow you to design additional hypothesis-directed experiments.
Barcoding libraries contain plasmids with unique, semi-random or random sequences that can be used for applications like lineage tracing, drug screening, and gene function studies. In gRNA libraries, each plasmid contains a unique gene targeting sequence. Surface display libraries allow the ability to screen large, diverse libraries for specific binding properties without the need to purify individual proteins. Screening libraries (non-CRISPR) can be used for different types of high-throughput experiments, including studying regulatory sequences, screening certain types of proteins (such as cell surface receptors or transcription factors), or observing the effects of mutagenesis of a single gene.
As you might have guessed from their name, pooled libraries are supplied as a mixed population of plasmids in a single tube.
Types of Pooled Libraries
Barcoding
Barcode libraries contain large numbers of plasmids, each with a short, randomized, unique nucleotide sequence, or barcode, used to mark individual cells. These barcodes will then be inherited by any daughter cells, making barcode libraries ideal for lineage tracing. Barcode libraries can be used for clonal fitness and competition assays, measuring bottlenecks after treatments, deconvolution of pooled perturbations, and linking perturbations to single-cell phenotypes. These libraries are also commonly used to monitor the dynamics of heterogeneous tumors.
Advantages
- Extremely high resolution clonal tracking
- Low biological perturbation
Limitations
- No functional information on genes unless combined with perturbations
- Requires deep sequencing
- Must maintain high representation
CRISPR
CRISPR is a useful tool for genetic screening experiments, due to the relative ease of designing gRNAs and the ability to modify virtually any genetic locus. CRISPR pooled libraries deliver many gRNAs at once, targeting many genes in one experiment. These libraries may be designed to target all the genes in the genome of an organism or a subset of genes, such as those involved in a certain pathway or biological process. In a CRISPR screening experiment, target cells are treated with the pooled library to create a population of mutant cells that are then screened for a phenotype of interest. Screening experiments using a pooled CRISPR library are far more complex than using CRISPR to modify a single genomic locus. CRISPR pooled libraries are also useful for loss-of-function screens, gain-of-function screens, enhancer/promoter mapping, and variant effect mapping studies. You can learn more about CRISPR tools in our CRISPR Guide.
There are multiple types of CRISPR libraries:
- Knockout: CRISPR knockout libraries are designed to create insertions or deletions in targeted genes across the genome, rendering them nonfunctional.
- Activation: CRISPR activation libraries direct an inactive Cas enzyme fused to a transcriptional activator to target genes or regulatory regions, where it stimulates transcription by recruiting activators or modifying local chromatin.
- Inhibition: CRISPR inhibition libraries direct an inactive Cas enzyme (alone or fused to a transcriptional repressor) to target genes, where it represses expression by blocking transcription initiation or modifying local chromatin.
- Base Edit: CRISPR base edit libraries use gRNAs to guide base editors to convert one base to another.
- Prime Edit: CRISPR prime edit libraries use prime editing guide RNAs (pegRNAs) to target genes for creating insertions, deletions, and base edits.
- Other: CRISPR libraries have also been designed for other specific purposes, such as barcoding or targeting RNA.
Advantages
- Strong, specific loss of function (knockout) or gain-of-function (activation) effects
- Genome-wide coverage possible
- Well-validated libraries
- Compatible with pooled next generation sequencing (NGS) or single-cell readouts
Limitations
- Double-stranded break toxicity (knockout)
- Chromatin-dependent efficiency (activation/inhibition)
- Essential genes may complicate knockout screens
- Requires Cas machinery, which may need to be expressed separately
- Large number of cells required to ensure adequate representation of every guide
Screening
Screening libraries (non-CRISPR) can be used for many different high-throughput experiments. They are useful for probing cellular pathways and mechanisms in gain- or loss-of-function screens, in vivo mutagenesis, pathway mapping, and drug resistance studies.
Advantages
- Broad range of perturbation methods
- Can target systems incompatible with CRISPR
- Can include gain- or loss-of-function
Limitations
- Efficiency and specificity vary by library type
- Often specific to a certain use or gene
Surface Display
Surface display libraries use a genetic engineering technique that physically links a protein's function (phenotype) to the gene that encodes it (genotype). This is achieved by fusing the gene of interest with a gene encoding for an anchoring protein. The resulting fusion protein is then expressed on the surface of a host cell or virus, such as bacteriophages or yeast.
Surface display libraries are useful for epitope mapping, protein engineering, and ligand discovery. The key advantage of this approach is the ability to screen large, diverse libraries for specific binding properties without the need to purify individual proteins. Any successful candidate can be readily identified by recovering and sequencing its associated genetic material.
Commonly used surface display platforms include:
- Phage Display: In this system, a peptide or protein is fused to a bacteriophage coat protein. The resulting phages can be screened for binding affinity to a target molecule.
- Yeast Display: In this system, proteins are displayed on the surface of Saccharomyces cerevisiae, often by fusion to the Aga2p protein. This platform supports the correct folding and post-translational modification of complex eukaryotic proteins, such as antibodies.
Advantages
- Explore vast sequence diversity
- Can map epitopes or binding partners
- Supports directed evolution
Limitations
- Protein-specific, may require specialized systems
- Limited to extracellular interactions or display-compatible proteins
cDNA/ORF Overexpression
cDNA libraries are created from an organism’s actively transcribed mRNA, as opposed to genomic libraries, which are created from genomic DNA. Since cDNA libraries lack introns and other nontranscribed sequences, they are smaller and easier to use than genomic libraries, but they do not contain as much regulatory information. To make a cDNA library, cDNA is synthesized, ligated to adapters, and then cloned into a given vector. They are useful for gain-of-function screens, pathway discovery, and functional complementation.
Advantages
- Strong and uniform overexpression
- Independent of chromatin accessibility
- Direct gain-of-function phenotypes
Limitations
- Non-physiological expression
- Cannot capture regulatory/isoform complexity
shRNA/RNAi
shRNA libraries permit reversible loss-of-function screening to elucidate which genes are involved in a phenotype. They are also useful for drug resistance screens and noncoding RNA studies. Each library contains multiple shRNA sequences for each target gene – a true positive hit (a particular target gene) in an shRNA screen should show consistent results from the multiple shRNAs that target it. shRNA libraries may also be barcoded to allow for easy identification of the shRNA a given cell carries. Addgene currently does not distribute any shRNA libraries, but previously distributed the DECIPHER libraries.
Advantages
- Works in cells incompatible with CRISPR
- Avoids double-strand break toxicity
- Can target noncoding RNAs
- Partial knockdown can model haploinsufficiency
Limitations
- Variable knockdown efficiency
- Partial versus complete loss-of-function
Amplifying and Using the Library
Once you receive your library from Addgene, check the library information to see if it should be amplified before you conduct your screen. If you need to amplify the library, please refer to the depositor’s protocol for the best results. If you have any questions about the library protocols, feel free to contact us at [email protected] for assistance.
For some libraries, plasmid DNA can be delivered directly to the cells of interest. With others, notably pooled lentiviral plasmid libraries, the plasmids must first be used to make viral vectors. This pooled viral vector library is subsequently used to deliver the plasmids to the cells of interest. In either case, NGS of the amplified library DNA is recommended to verify that the library contains all of the expected plasmids — an incomplete library may lead to false positives or false negatives in later experiments, and can also negatively affect data reproducibility.
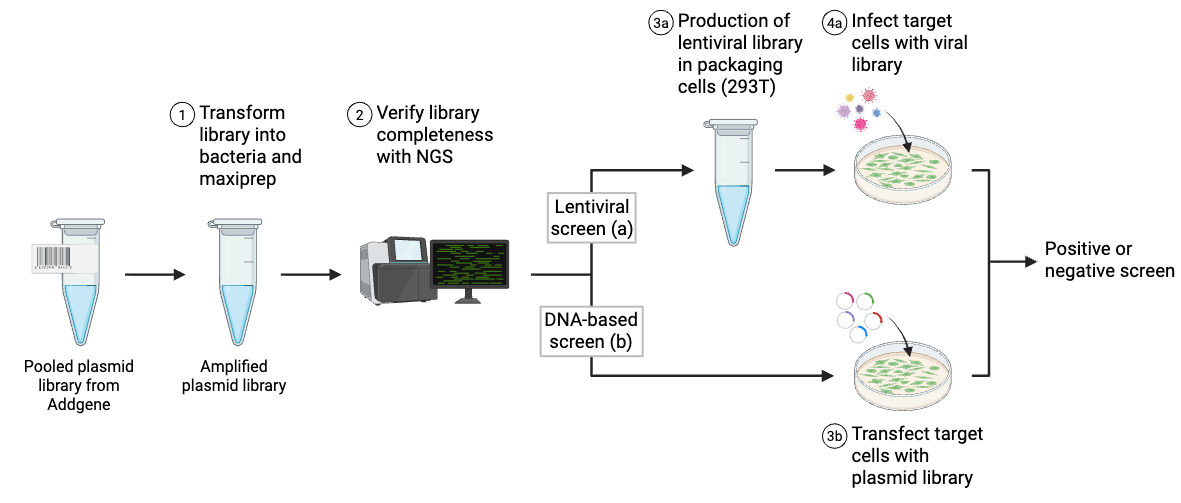
Types of Pooled Library Screens
In pooled lentiviral library screens, cells are transduced at a very low multiplicity of infection (MOI) to increase the odds that an transduced cell receives only one plasmid. To make sure that every plasmid is adequately represented in the population, you’ll need to transduce many more cells than the number of plasmids present in the library – for example, the Moffat lab uses 200x more cells than plasmids in the TKO library for their screen. Using large numbers of cells minimizes the effects of random chance that could lead to false positive or negative results – for a given plasmid, think of each cell carrying it as being a biological replicate for that plasmid.
Library screens can be divided into two types: positive screens and negative screens. Both types of screen employ a selection method relevant to the phenotype being studied. An example of a published selection mechanism for lentiviral CRISPR libraries is resistance to the nucleotide analog 6-thioguanine. Because NGS is required to accurately identify which library elements are affected by selection, access to this technology is essential for any pooled library screen.
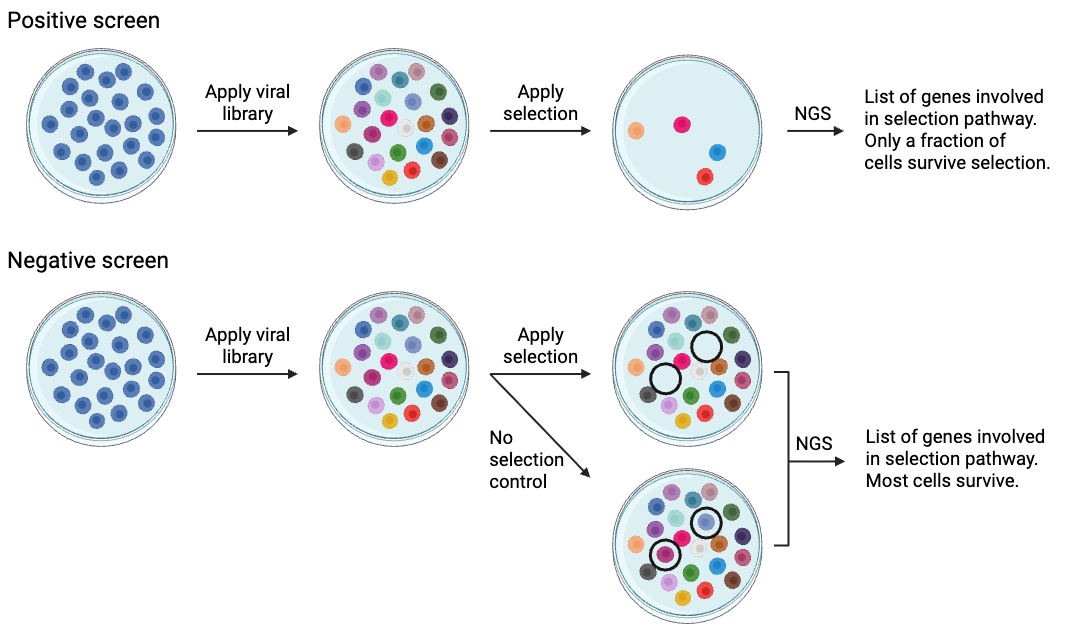
Both positive and negative screens follow the same basic process. Before applying to cells, the library must be amplified (electroporation and maxiprep) and packaged into lentiviral vectors (if delivering by viral vectors). The screens diverge at a few important points, including when to apply selection, when to sequence, and how to analyze the results.
Positive screen:
- Apply library
- Apply selection, most cells die or do not pass selection (in the case of a reporter)
- Sort selected cells
- Sequence those cells that are selected
- Get a list of genes involved in the selection
In a positive screen, the goal is to identify cells that survive selection. The selective pressure must be strong enough that most of the cells die, removing their plasmids from the population, and only a small fraction survive. After the surviving cells are collected, their plasmids are PCR-amplified and sequenced using NGS to identify their cDNA or target gene. Positive selection screens are generally very robust, and tens of thousands of genes that were initially present may be narrowed down to several hundred or fewer.
Negative screen:
- Apply library
- Perform NGS on a control sample (no selection)
- Apply selection; most cells will survive
- Perform NGS on the surviving cells
- Compare NGS results between experimental and control cells
- Generate a list of library plasmids that disappear with addition of the selection mechanism
Negative screens are a little trickier than positive screens. In a negative screen, the goal is to identify those cells that do not survive the selection mechanism. You’ll transduce two sets of cells and subject one set to selection while the other serves as a untreated control. These two populations are then sequenced using NGS to determine which plasmids have been depleted by selection. In a CRISPR screen, negative screens are often used to identify genes that are essential for growth/survival under certain conditions.
Next-Generation Sequencing
NGS allows for parallel sequencing of millions of DNA fragments simultaneously in a single reaction. As discussed above, access to this technology is crucial for conducting pooled library screens. Addgene does not offer NGS services. If you need assistance perfroming NGS, we recommend reaching out to your core facility, neighboring labs, or a commercial company near you.
Resources
Addgene Pooled Library Resources
- Learn about Pooled Library Handling and Storage.
- Deposit a Pooled Library and create a custom Pooled Library Amplification protocol.
- See our Pooled Library Amplification Protocol for Addgene's generalized library amplification protocol.
- Read our CRISPR guide for more information on genome-wide screening with CRISPR.
- Browse our Viral Vector resources.
Addgene Blog
- Genome-wide Screening Using CRISPR/Cas9
- Lentiviral CRISPR Libraries Enable Genome-Scale, Knockout Screening
- Pooled CRISPR Libraries Offer Genome-Wide Control for Large-Scale Functional Screens
- New Tool for Lineage Tracing: The ClonTracer Library
- NGS Quality Control for Pooled Libraries
- A Tour of Addgene's Most Popular Pooled Libraries
- CRISPR pooled library posts
Content last reviewed: 5 January 2026


