We narrowed to 1,329 results for: 5-Oct
-
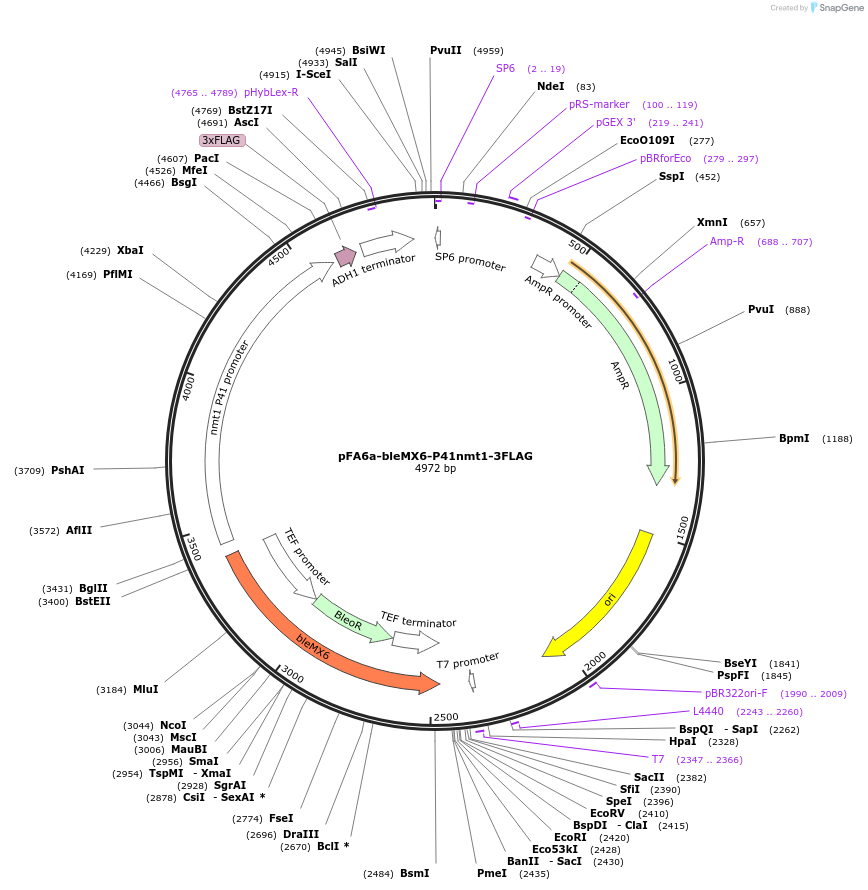
Plasmid#19352DepositorInsertbleMX6
ExpressionYeastAvailable SinceSept. 25, 2008AvailabilityAcademic Institutions and Nonprofits only -
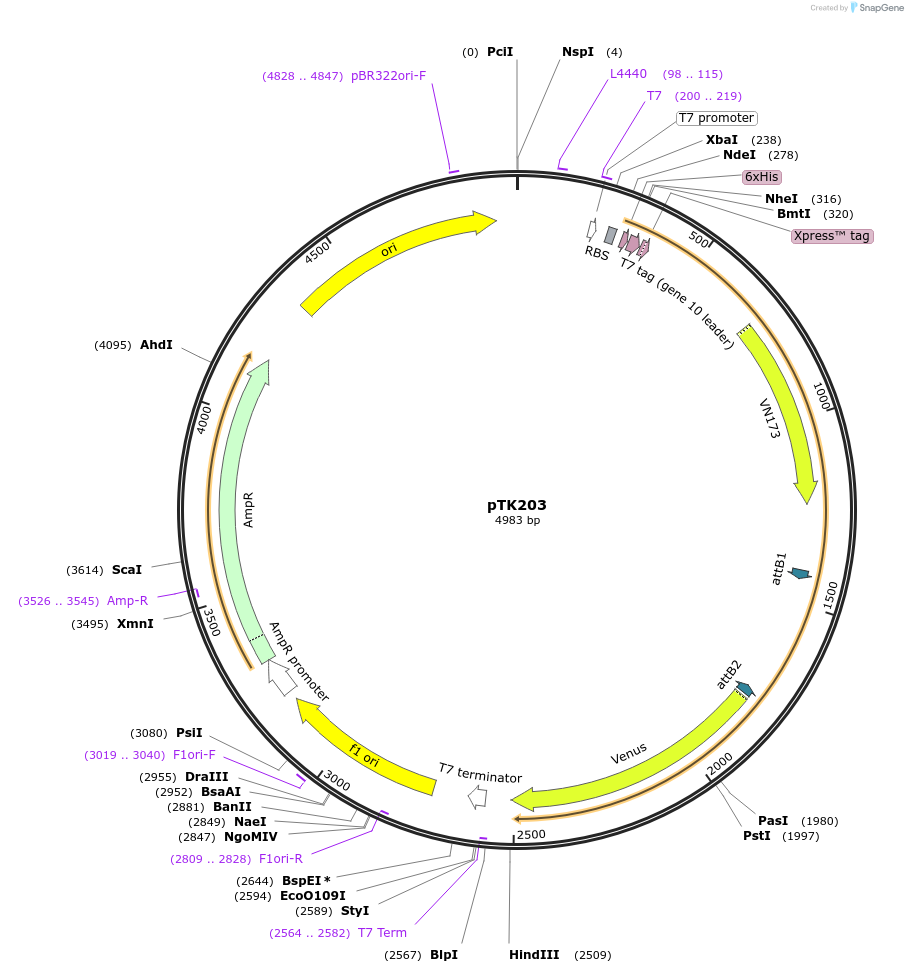
pTK203
Plasmid#13536DepositorInsertFLIPW-TCTY
TagsSee mapExpressionBacterialAvailable SinceJuly 27, 2007AvailabilityAcademic Institutions and Nonprofits only -
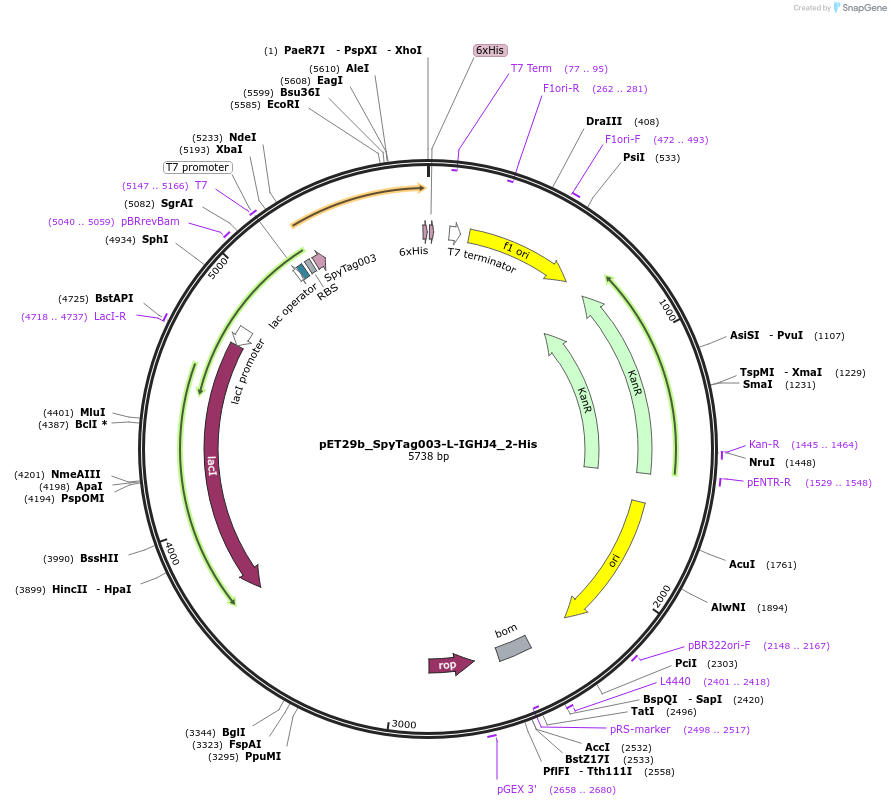
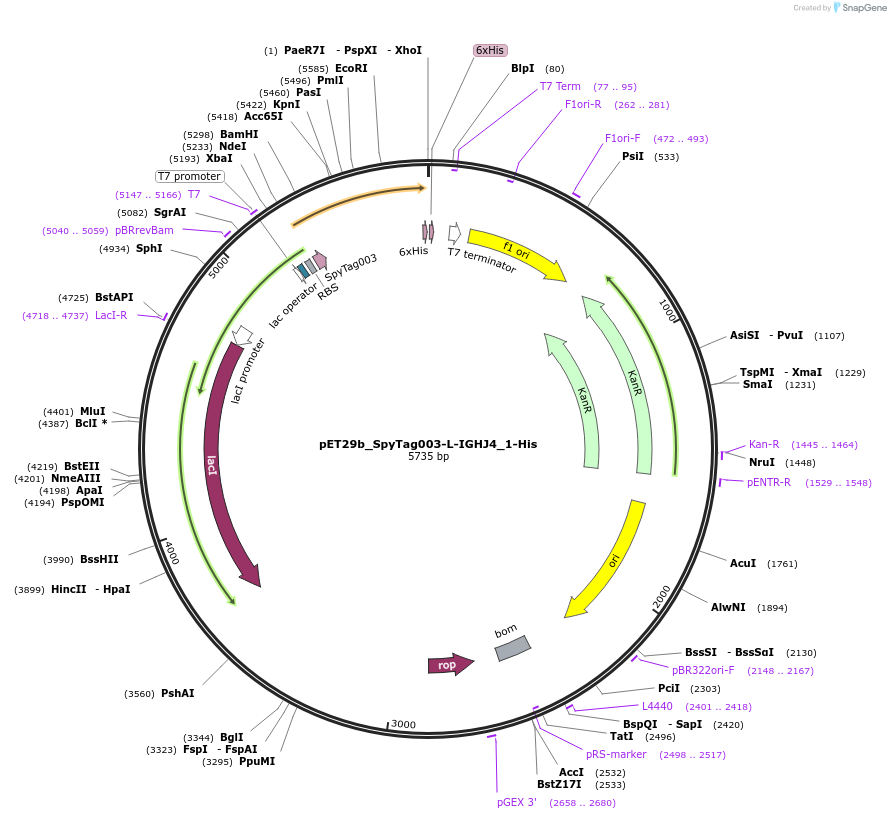
pET29b_SpyTag003-L-IGHJ4_2-His
Plasmid#233335PurposeTo express nanobody IGHJ4_2 with N terminal SpyTagDepositorInsertSpyTag003-L-IGHJ4_2-His
ExpressionBacterialPromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
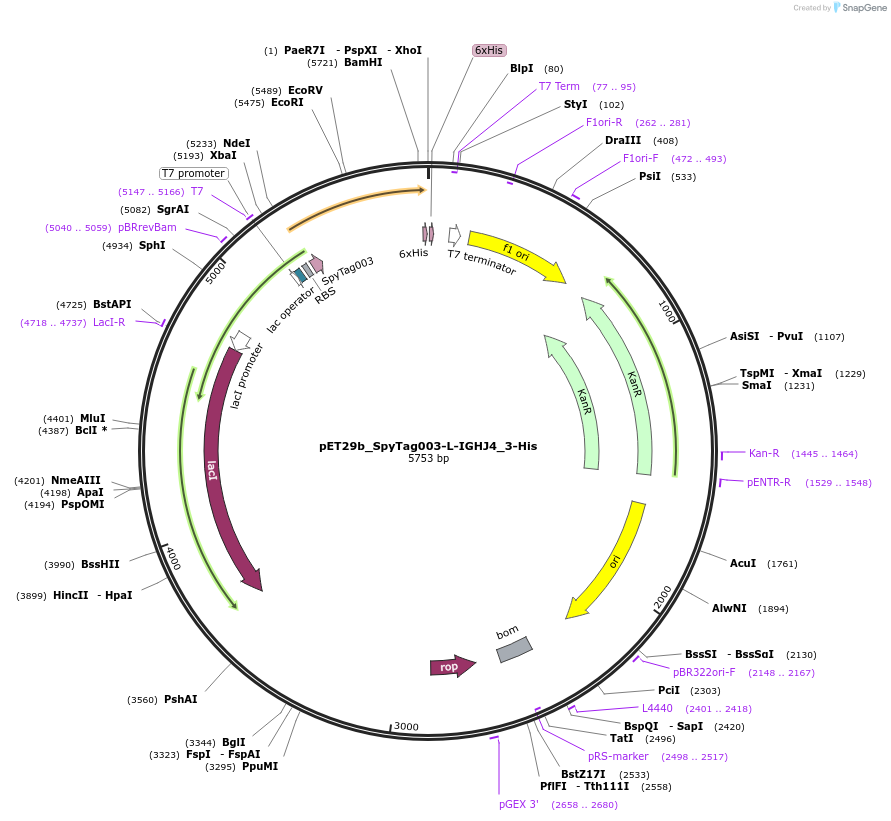
pET29b_SpyTag003-L-IGHJ4_3-His
Plasmid#233336PurposeTo express nanobody IGHJ4_3 with N terminal SpyTagDepositorInsertSpyTag003-L-IGHJ4_3-His
ExpressionBacterialPromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
pET29b_SpyTag003-L-IGHJ4_4-His
Plasmid#233337PurposeTo express nanobody IGHJ4_4 with N terminal SpyTagDepositorInsertSpyTag003-L-IGHJ4_4-His
ExpressionBacterialPromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
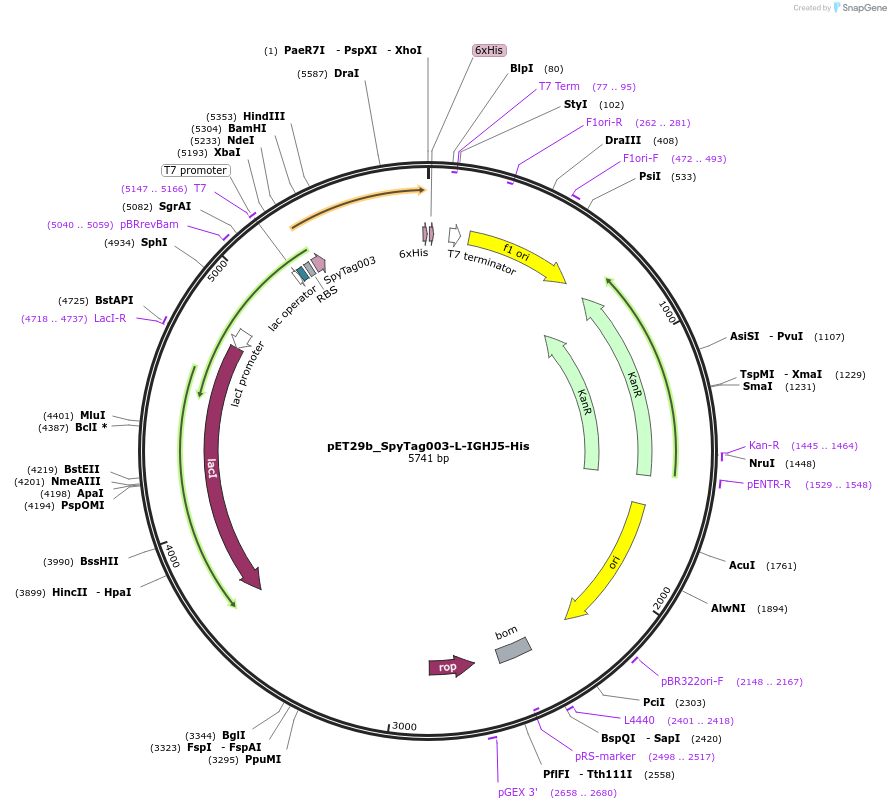
pET29b_SpyTag003-L-IGHJ5-His
Plasmid#233338PurposeTo express nanobody IGHJ5 with N terminal SpyTagDepositorInsertSpyTag003-L-IGHJ5-His
ExpressionBacterialPromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
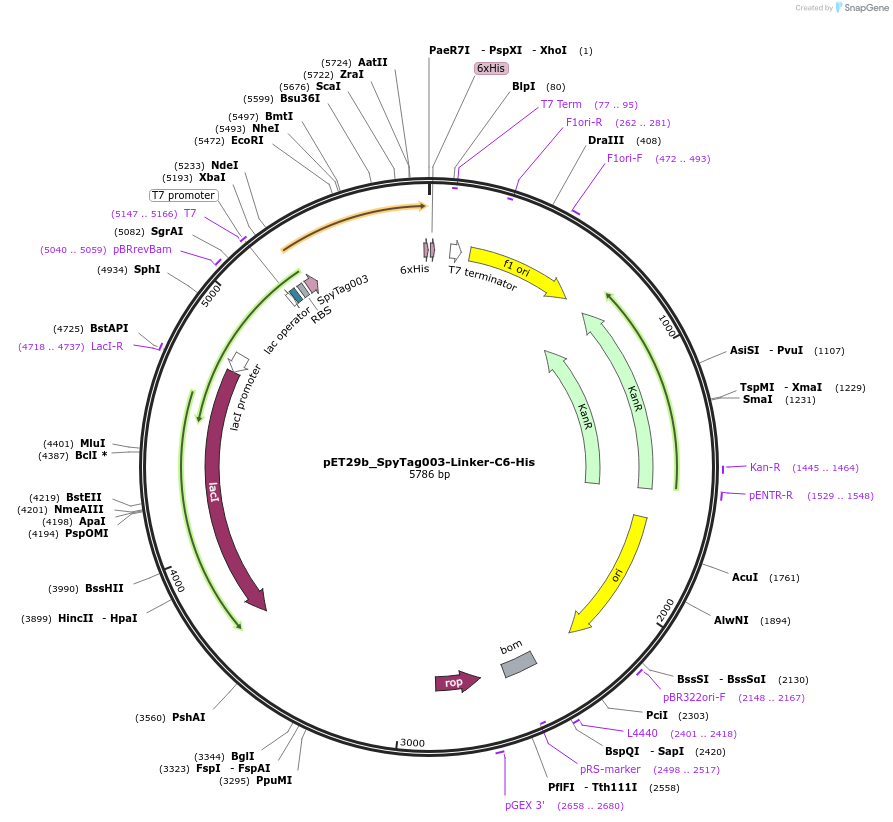
pET29b_SpyTag003-Linker-C6-His
Plasmid#233331PurposeTo express nanobody C6 with N terminal SpyTagDepositorInsertSpyTag003-Linker-C6-His
ExpressionBacterialPromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
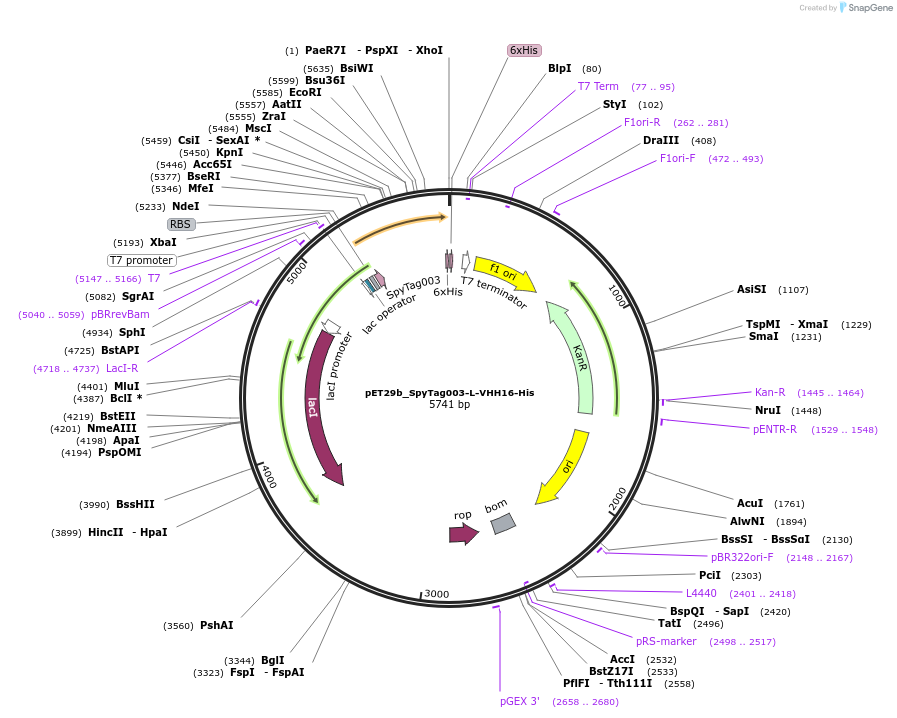
pET29b_SpyTag003-L-VHH16-His
Plasmid#233332PurposeTo express nanobody VHH16 with N terminal SpyTagDepositorInsertSpyTag003-L-VHH16-His
ExpressionBacterialPromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
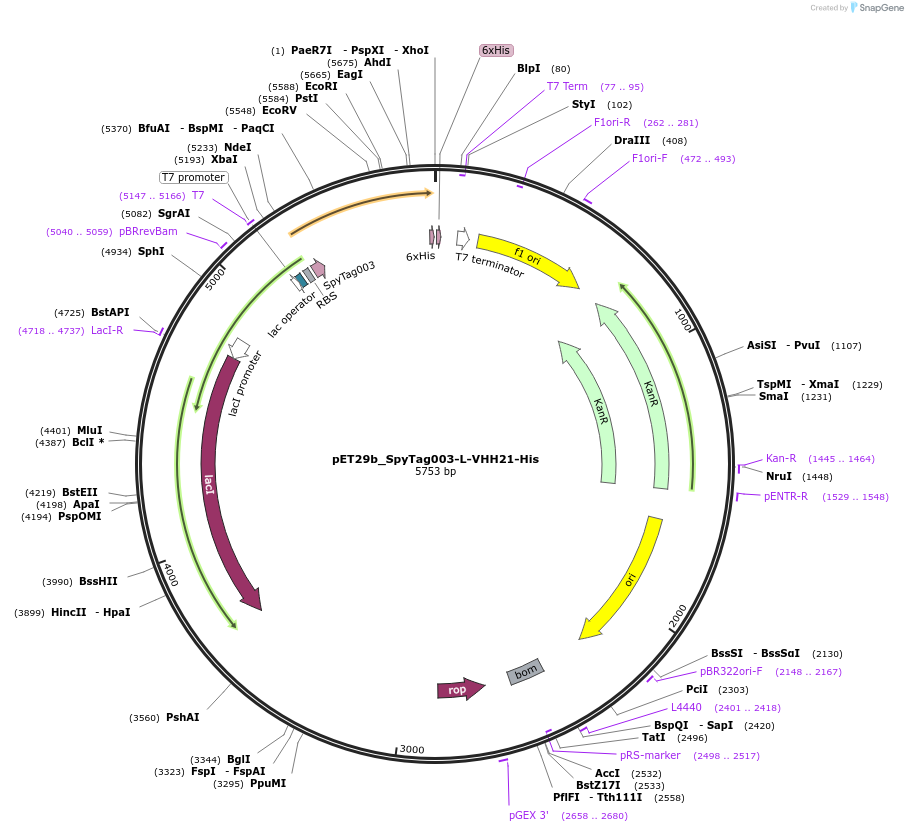
pET29b_SpyTag003-L-VHH21-His
Plasmid#233333PurposeTo express nanobody VHH21 with N terminal SpyTagDepositorInsertSpyTag003-L-VHH21-His
ExpressionBacterialPromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
pET29b_SpyTag003-L-IGHJ4_1-His
Plasmid#233334PurposeTo express nanobody IGHJ4_1 with N terminal SpyTagDepositorInsertSpyTag003-L-IGHJ4_1-His
ExpressionBacterialPromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
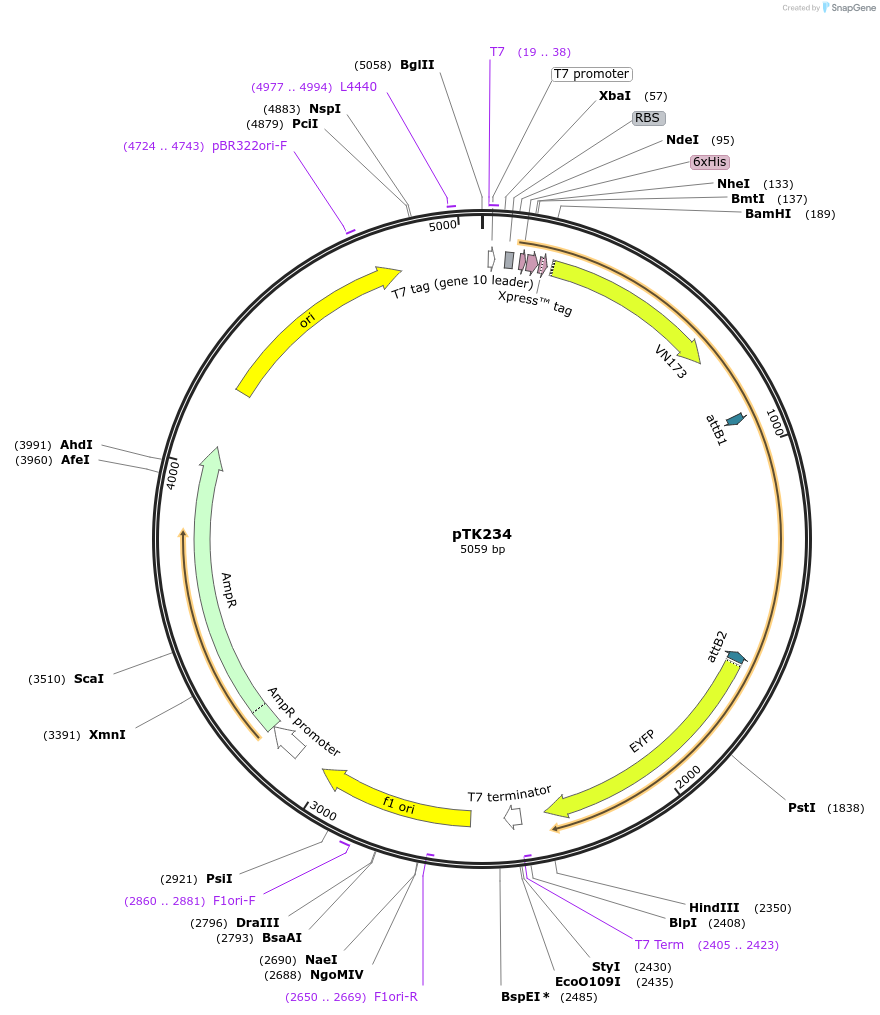
pTK234
Plasmid#13535DepositorInsertFLIPW-CTTY
TagsSee mapExpressionBacterialAvailable SinceJuly 27, 2007AvailabilityAcademic Institutions and Nonprofits only -
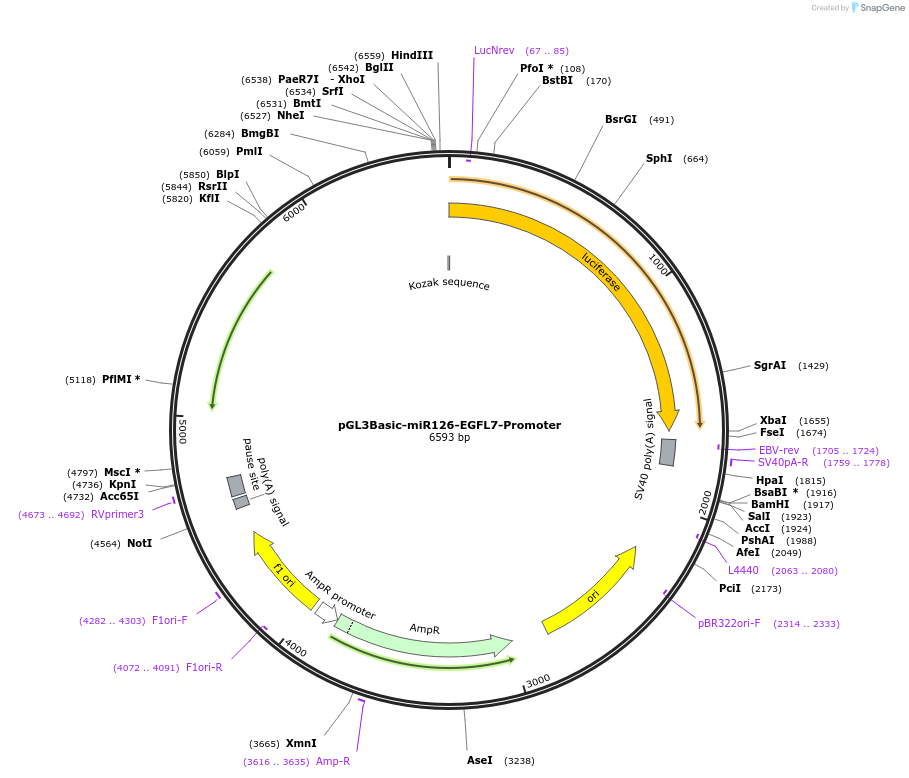
pGL3Basic-miR126-EGFL7-Promoter
Plasmid#32244DepositorUseLuciferaseExpressionMammalianMutationPromoter region 5' UTRPromoter5' UTRAvailable SinceSept. 19, 2011AvailabilityAcademic Institutions and Nonprofits only -
hChT pcDNA3
Plasmid#15766DepositorInserthemicholinium-3-sensitive choline transporter (SLC5A7 Human)
ExpressionMammalianAvailable SinceSept. 13, 2007AvailabilityAcademic Institutions and Nonprofits only -
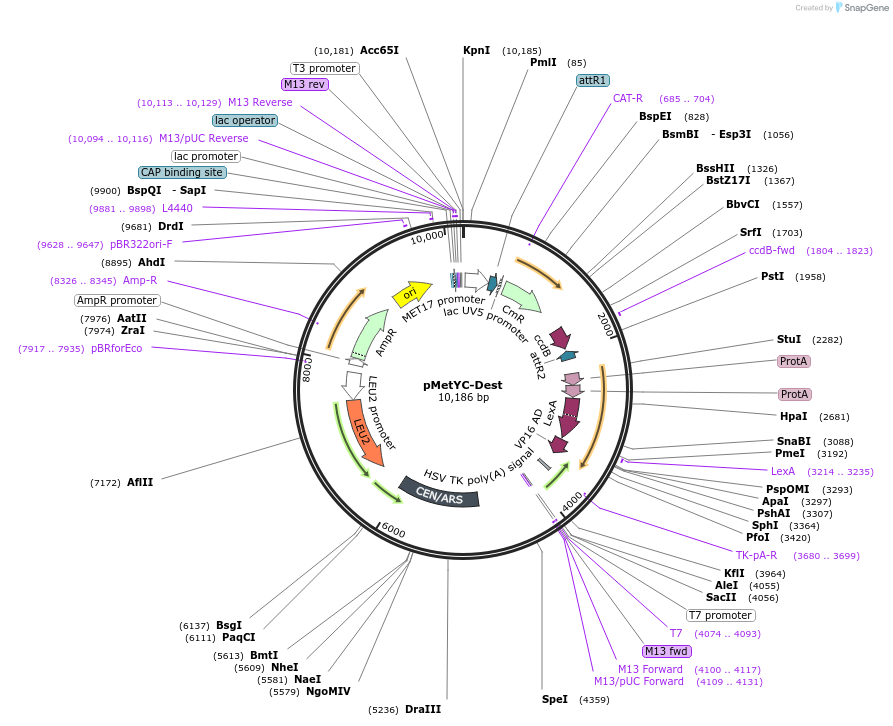
pMetYC-Dest
Plasmid#105081PurposeYeast mating-based Split Ubiquitin Assay, bait vector for Gateway cloning. Cub-PLV does not contain a start codon.DepositorTypeEmpty backboneTagsCub-PLVExpressionYeastAvailable SinceFeb. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
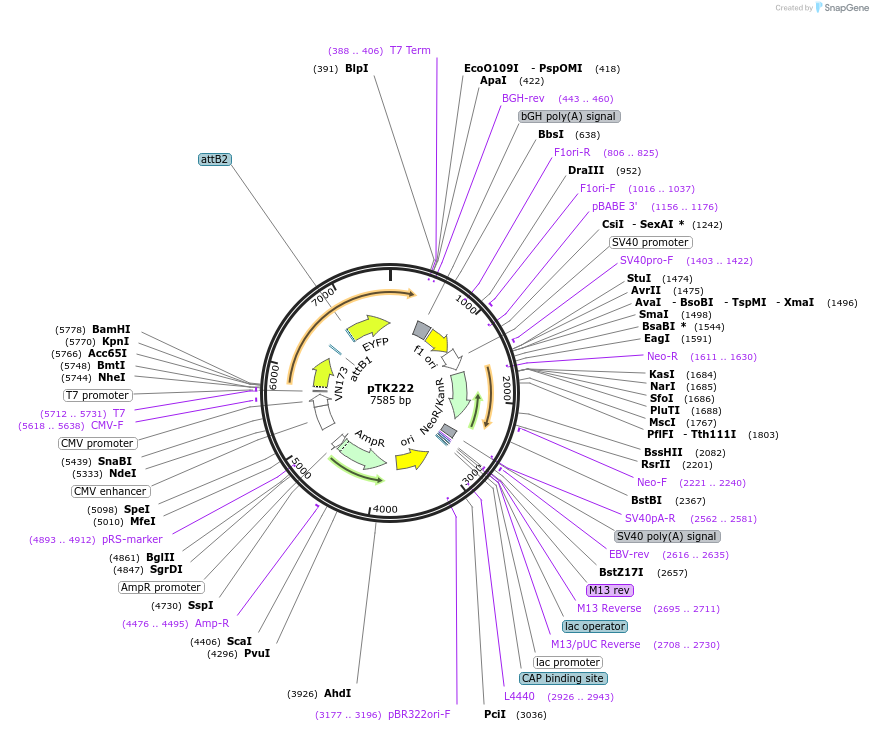
pTK222
Plasmid#13532DepositorInsertFLIPW-CTYT
TagsSee mapExpressionMammalianAvailable SinceJuly 27, 2007AvailabilityAcademic Institutions and Nonprofits only -
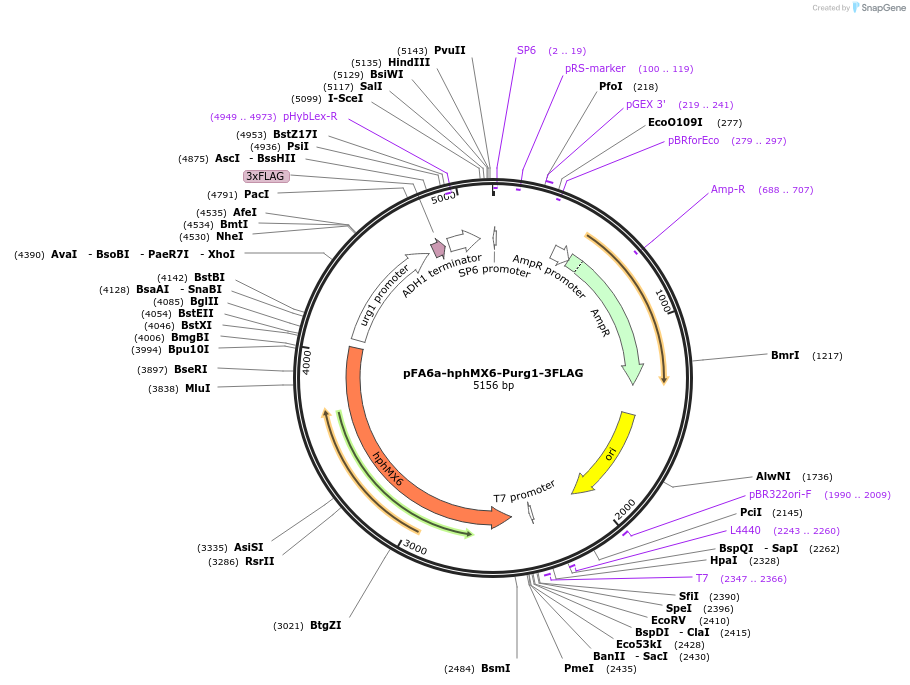
pFA6a-hphMX6-Purg1-3FLAG
Plasmid#19356DepositorInserturg1 promoter
ExpressionYeastAvailable SinceSept. 19, 2008AvailabilityAcademic Institutions and Nonprofits only -
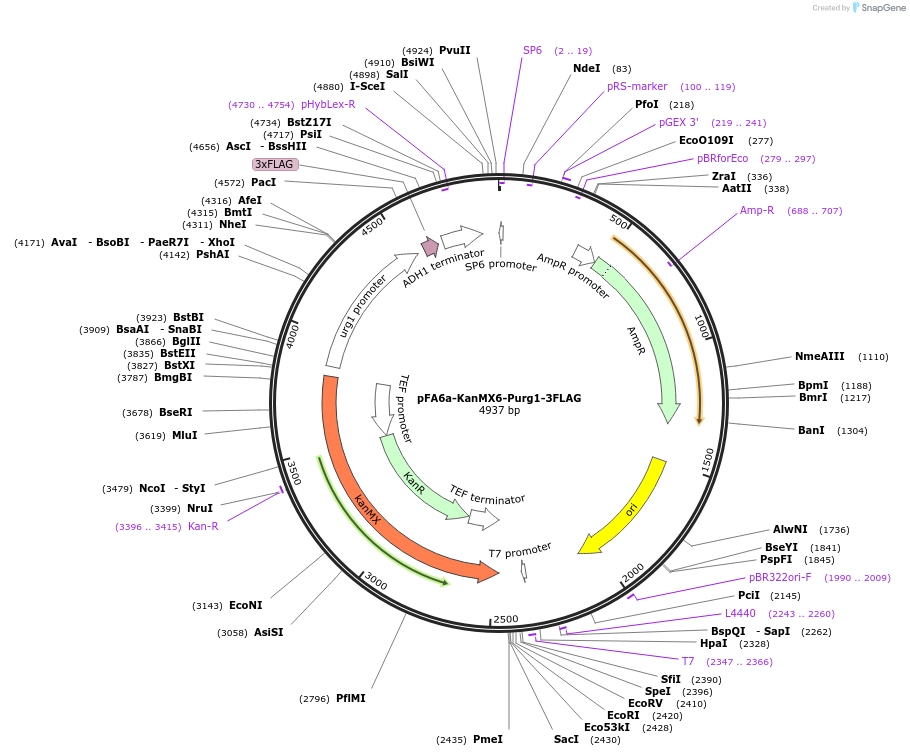
pFA6a-KanMX6-Purg1-3FLAG
Plasmid#19354DepositorInserturg1 promoter
ExpressionYeastAvailable SinceSept. 19, 2008AvailabilityAcademic Institutions and Nonprofits only -
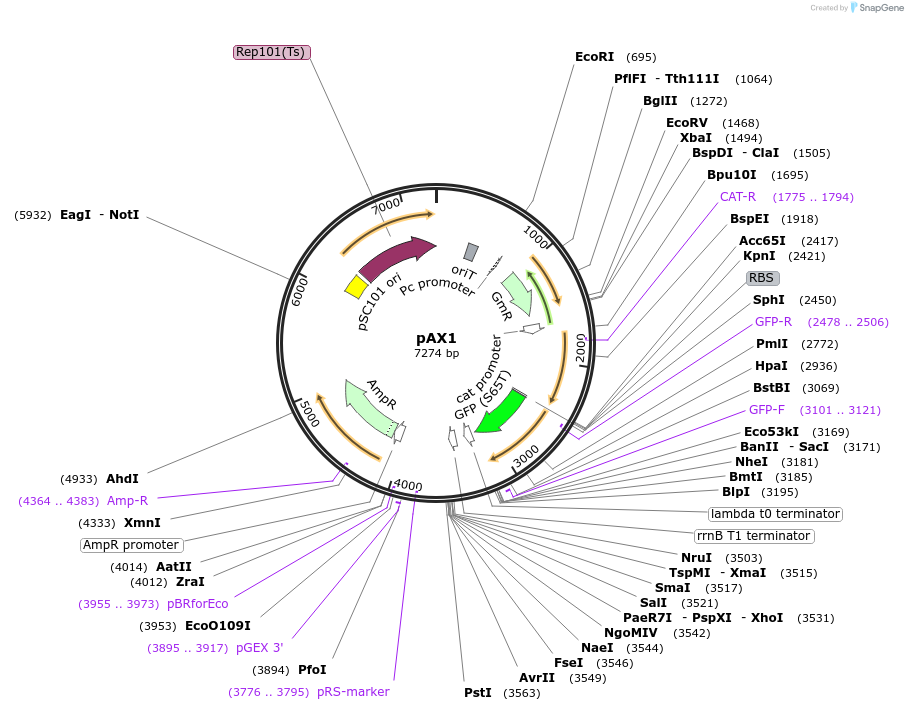
pAX1
Plasmid#117397Purposeallelic exchange vector with GFP merodiploid tracker and temperature-sensitive origin of replicationDepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialAvailable SinceMarch 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
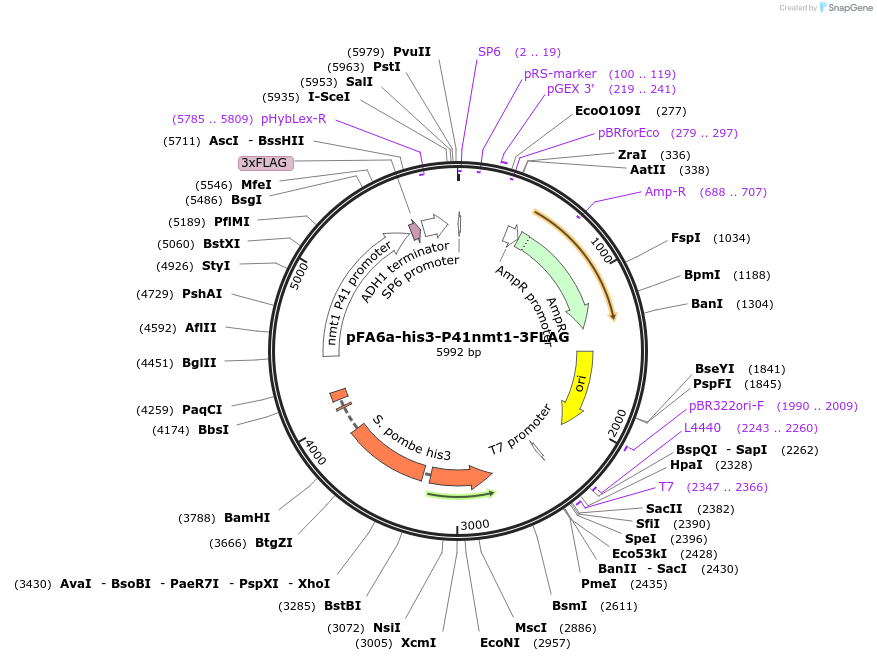
pFA6a-his3-P41nmt1-3FLAG
Plasmid#19340DepositorInserthis3 (his3 Fission Yeast)
ExpressionYeastAvailable SinceSept. 19, 2008AvailabilityAcademic Institutions and Nonprofits only -
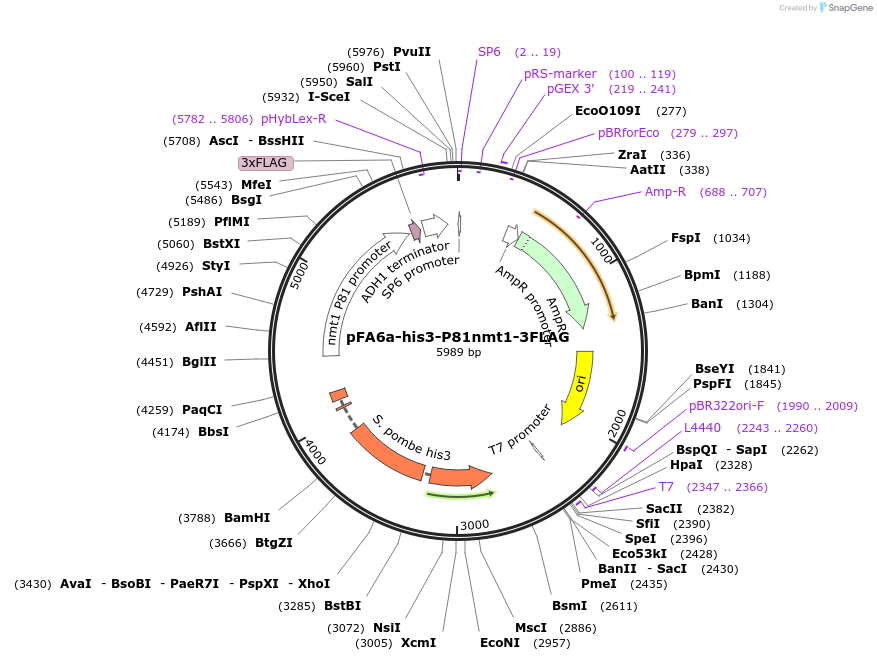
pFA6a-his3-P81nmt1-3FLAG
Plasmid#19341DepositorInserthis3 (his3 Fission Yeast)
ExpressionYeastAvailable SinceSept. 19, 2008AvailabilityAcademic Institutions and Nonprofits only