We narrowed to 2,507 results for: mia
-
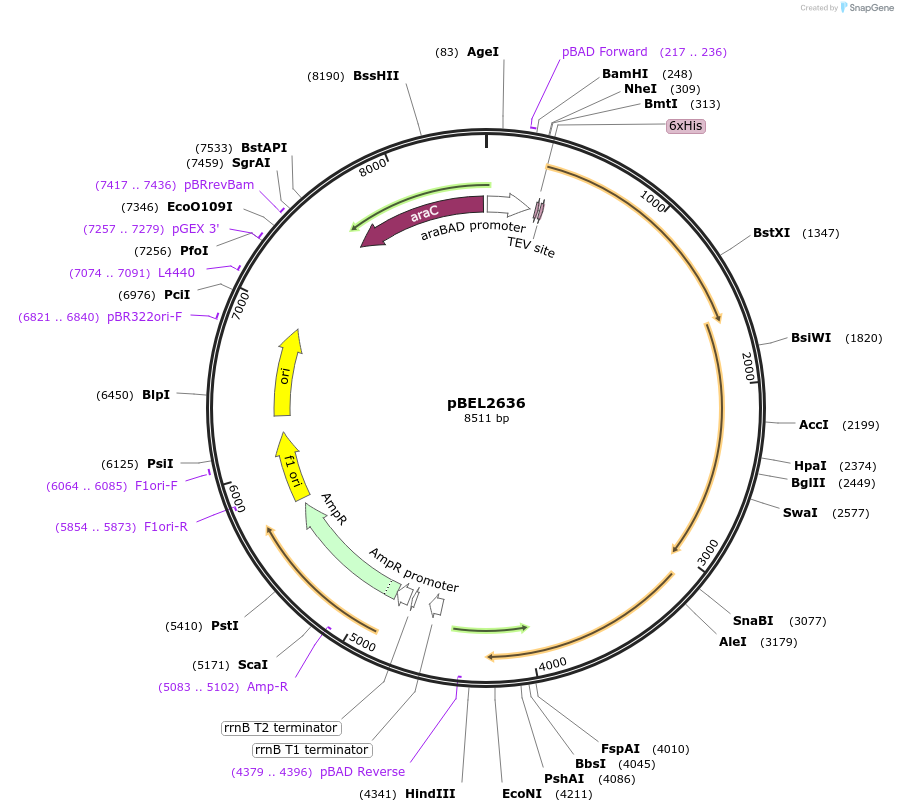
Plasmid#232500PurposeExpression plasmid of of LetA with LetB(F468B)DepositorTags6xHis2xQH-TEVExpressionBacterialMutationLetB F468AmberPromoteraraBADAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only
-
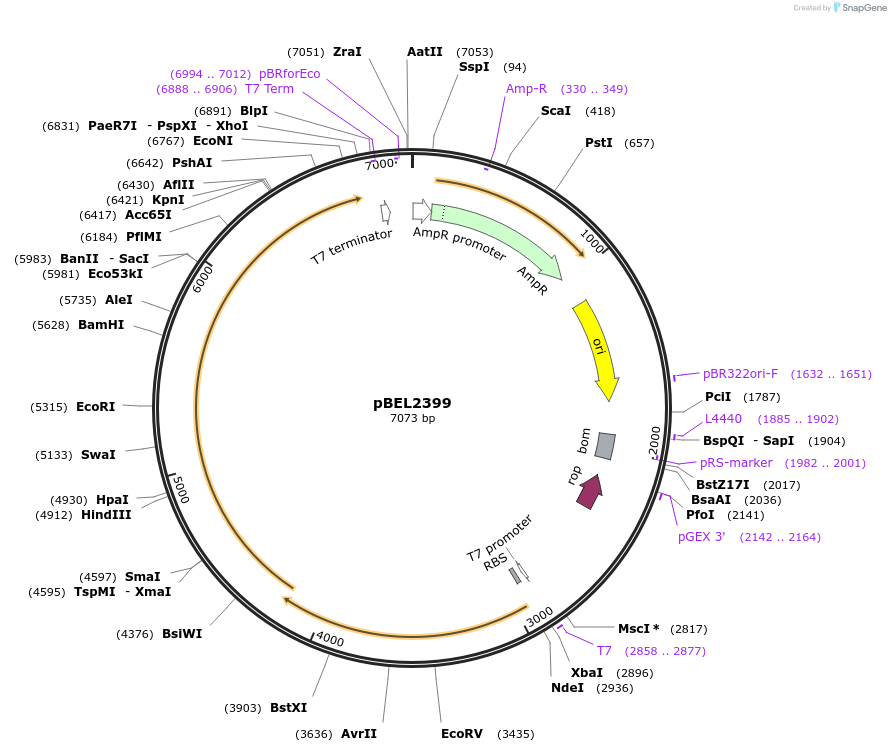
pBEL2399
Plasmid#232487PurposeGenetic complementation plasmid of LetA(C31A) with LetBDepositorAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
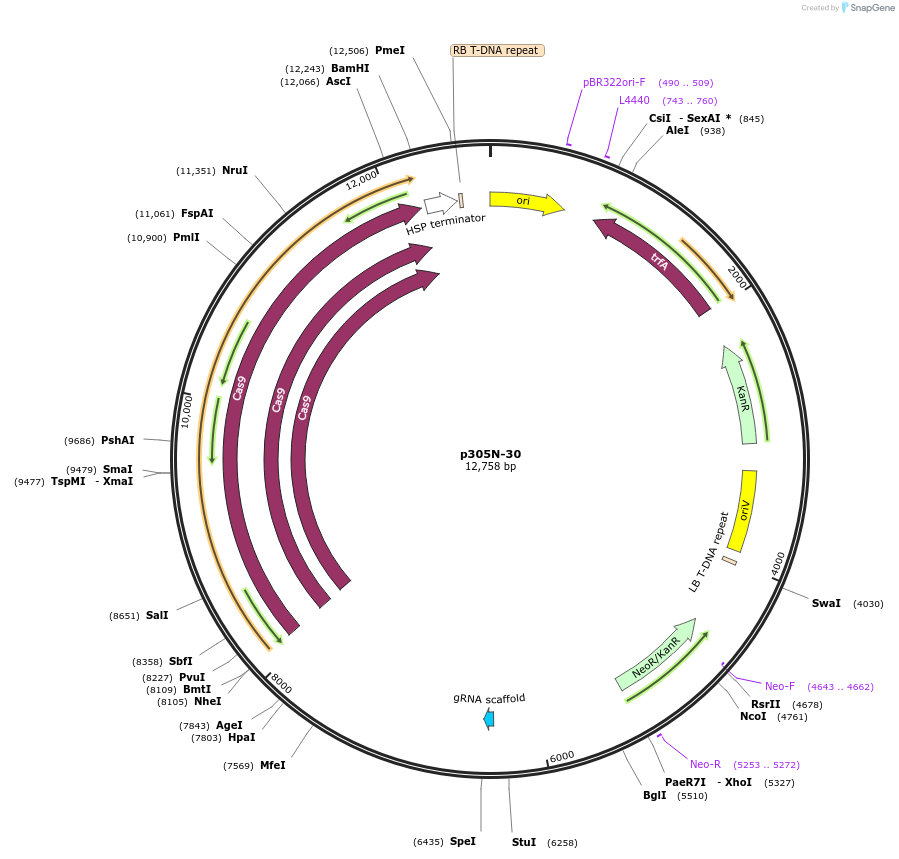
p305N-30
Plasmid#246313PurposeEvaluation of PtU6.2c3 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c3 promoter
UseCRISPRExpressionPlantMutationdeletion: -T (-29 from TSS) within TATAPromoterPtU6.2c3 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
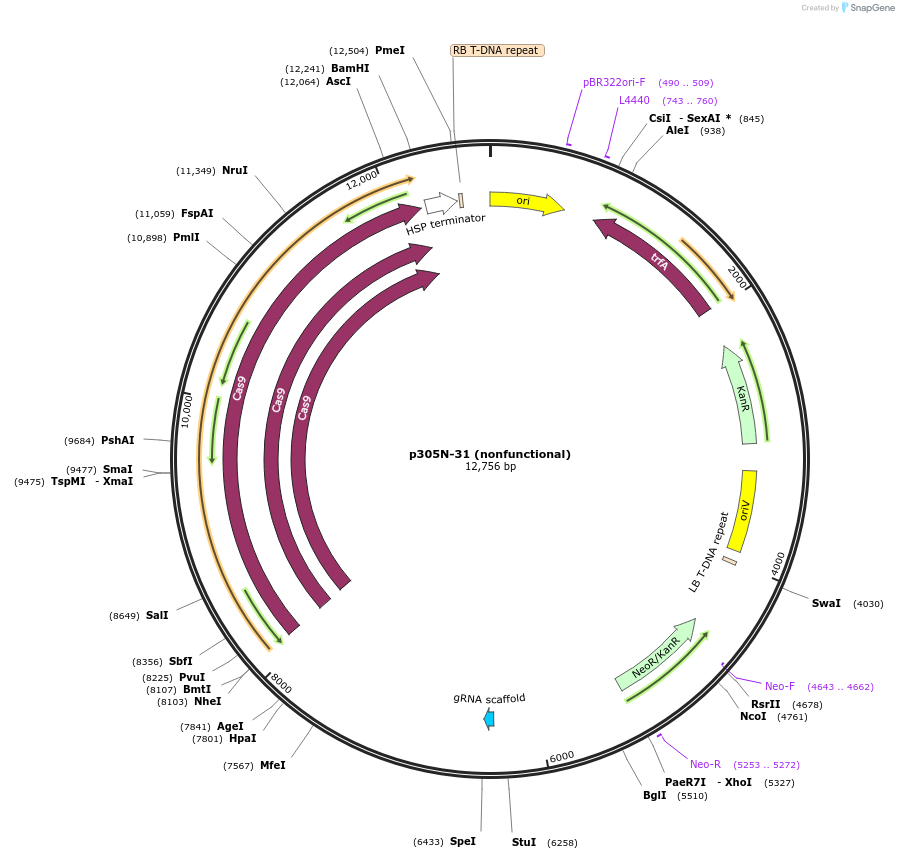
p305N-31 (nonfunctional)
Plasmid#246314PurposeEvaluation of PtU6.2c4 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c4 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationdeletions: -A (-41 from TSS), -T (-30), -T (-29);…PromoterPtU6.2c4 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
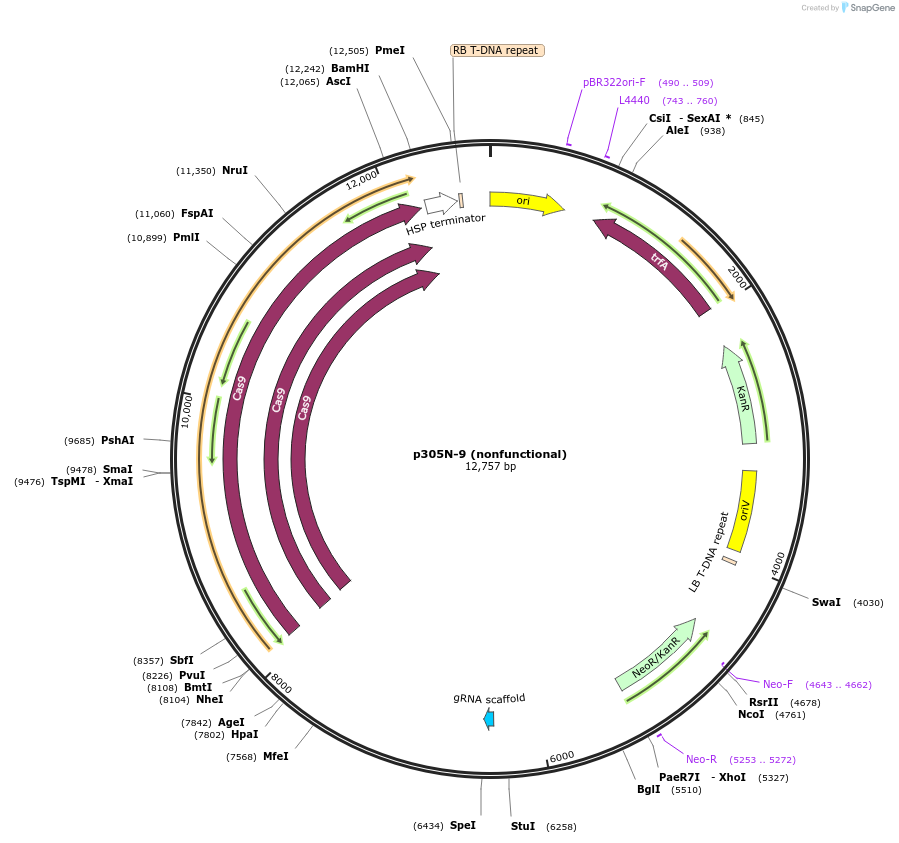
p305N-9 (nonfunctional)
Plasmid#246292PurposeEvaluation of AtU6.29c13 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.29c13 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationdeletions: -A (-58 from TSS), -T (-57) within USEPromoterAtU6.29c13 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
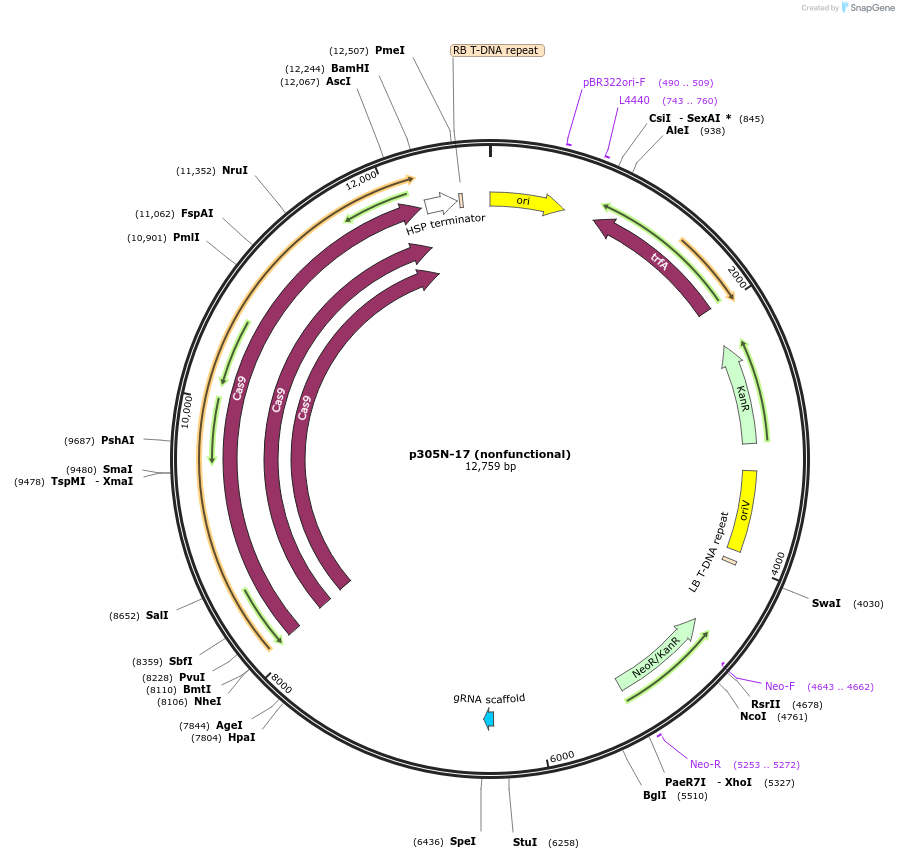
p305N-17 (nonfunctional)
Plasmid#246300PurposeEvaluation of HbU6.2m3 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertHbU6.2m3 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-T (-29 from TSS), G-to-A (-24)PromoterHbU6.2m3 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
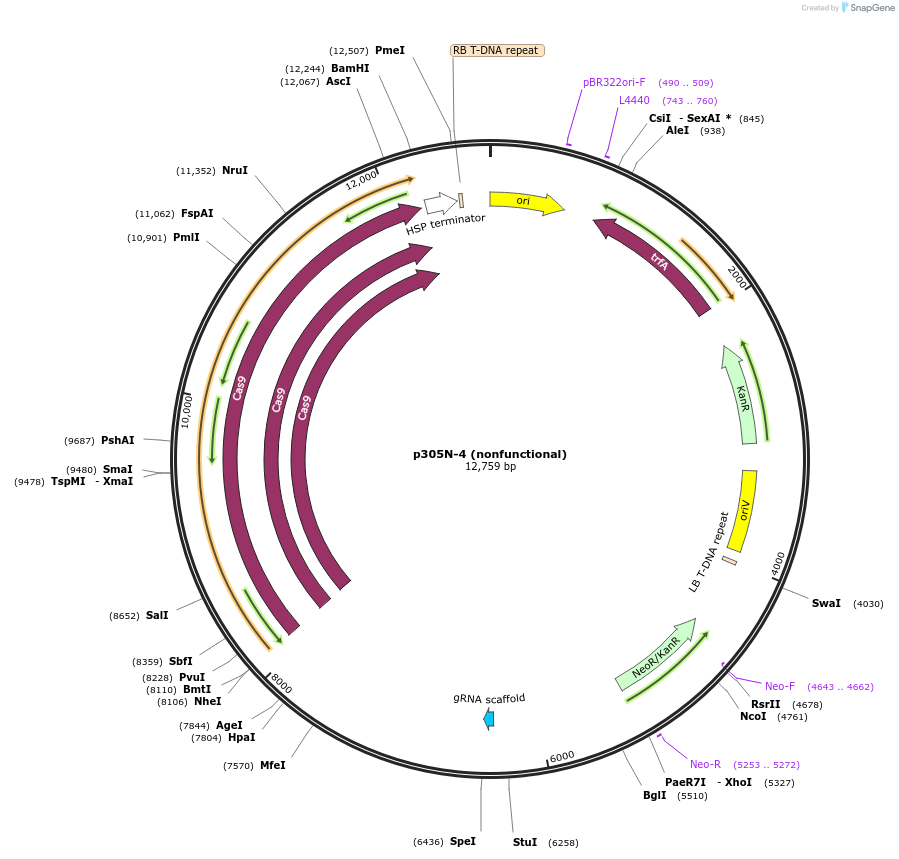
p305N-4 (nonfunctional)
Plasmid#246287PurposeEvaluation of AtU6.1c1 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.1c1 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationG-to-C at -15 from TSSPromoterAtU6.1c1 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
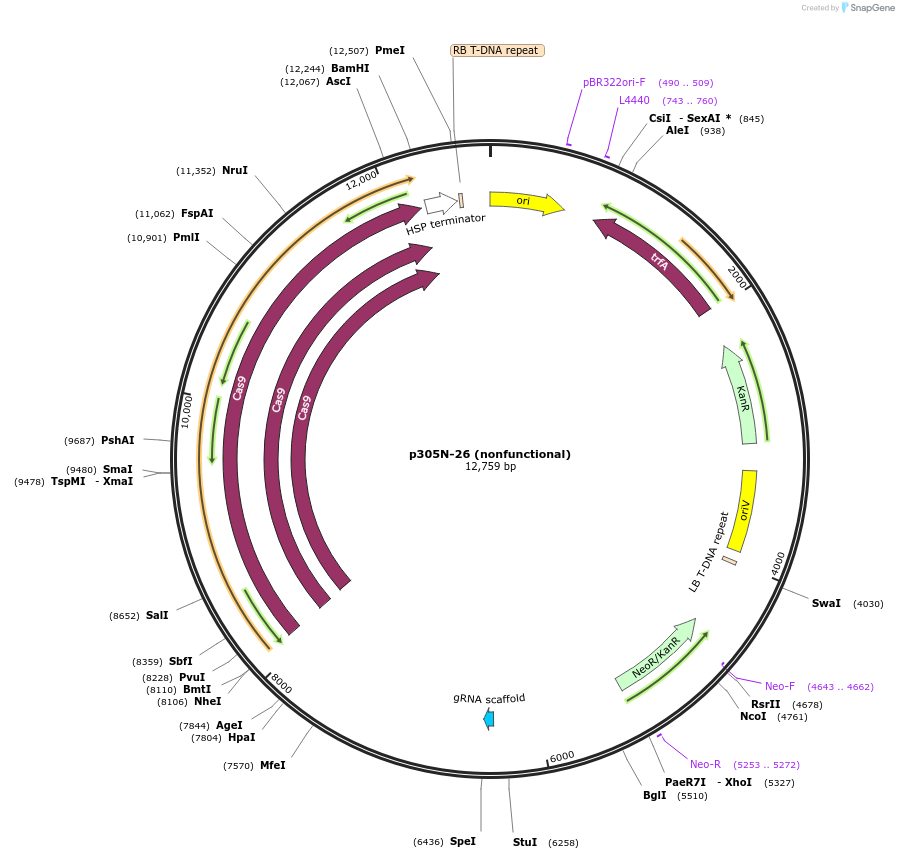
p305N-26 (nonfunctional)
Plasmid#246309PurposeEvaluation of MtU6.6m7 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertMtU6.6m7 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-A (-63 from TSS), T-to-G (-57)PromoterMtU6.6m7 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
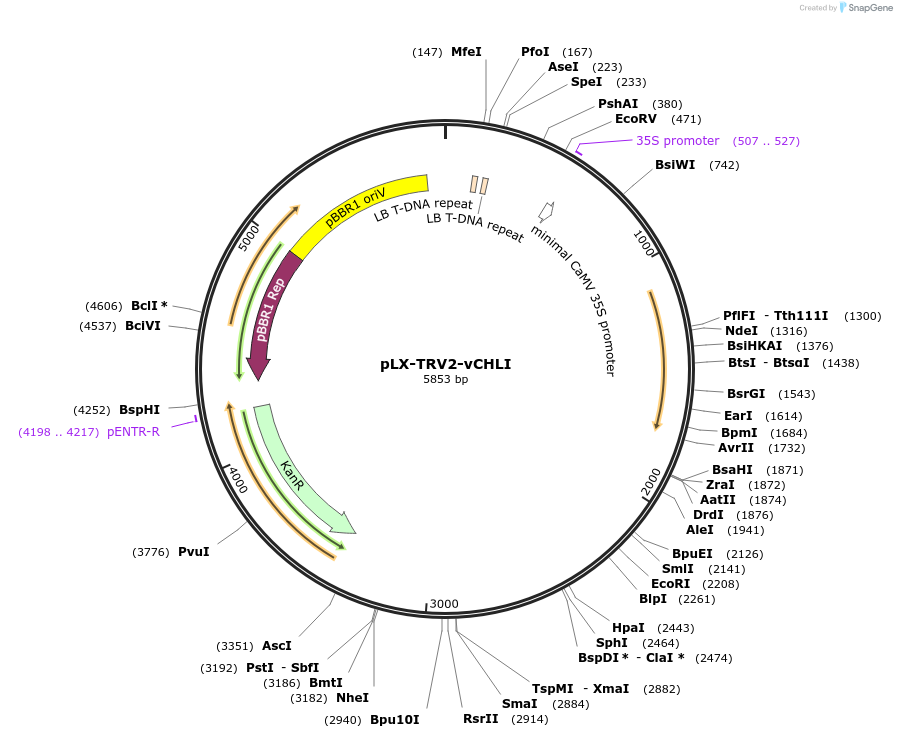
pLX-TRV2-vCHLI
Plasmid#239842PurposeDerivative of pLX-TRV2, a tobacco rattle virus (TRV) vector, designed for the expression of a 32-nt insert to silence CHLI homolog genes in Nicotiana benthamiana and Solanum aethiopicumDepositorInsertvCHLI
UseRNAiExpressionPlantAvailable SinceAug. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
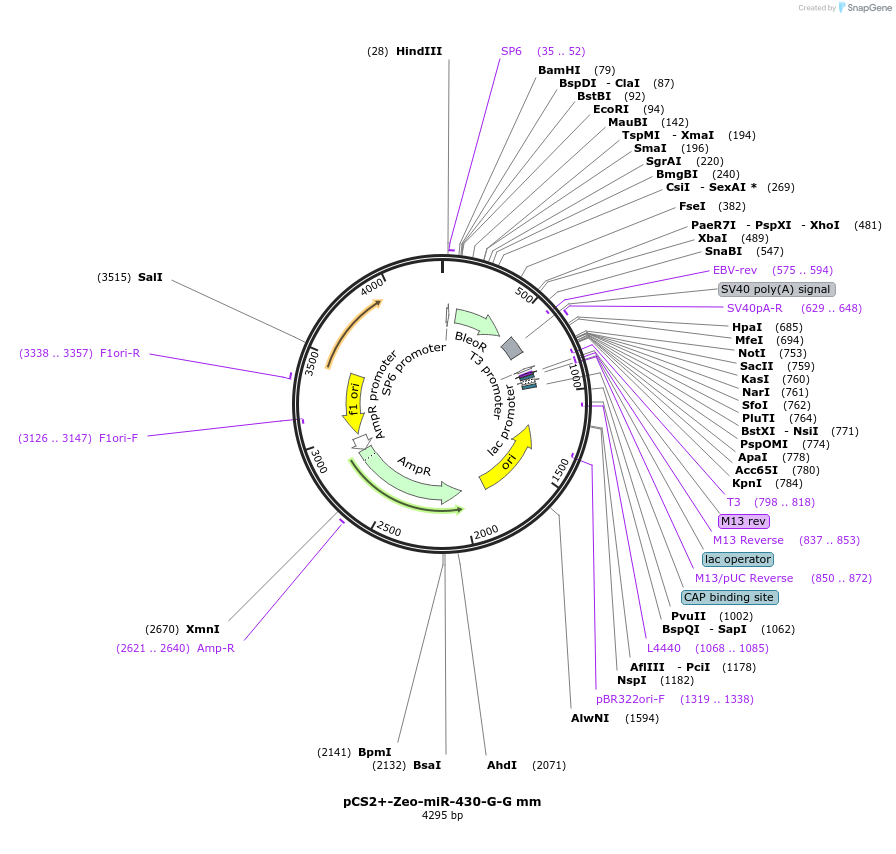
pCS2+-Zeo-miR-430-G-G mm
Plasmid#221728PurposeExpression of Zeocin resistance marker followed by miR-430 target site insertion with G-G mismatch at position 6 in zebrafish embryos.DepositorInsertBleoR-miR-430-G-G mm
UseFish expressionMutationG-G mismatch of miR-430 sitePromotersimian CMV IE94Available SinceDec. 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
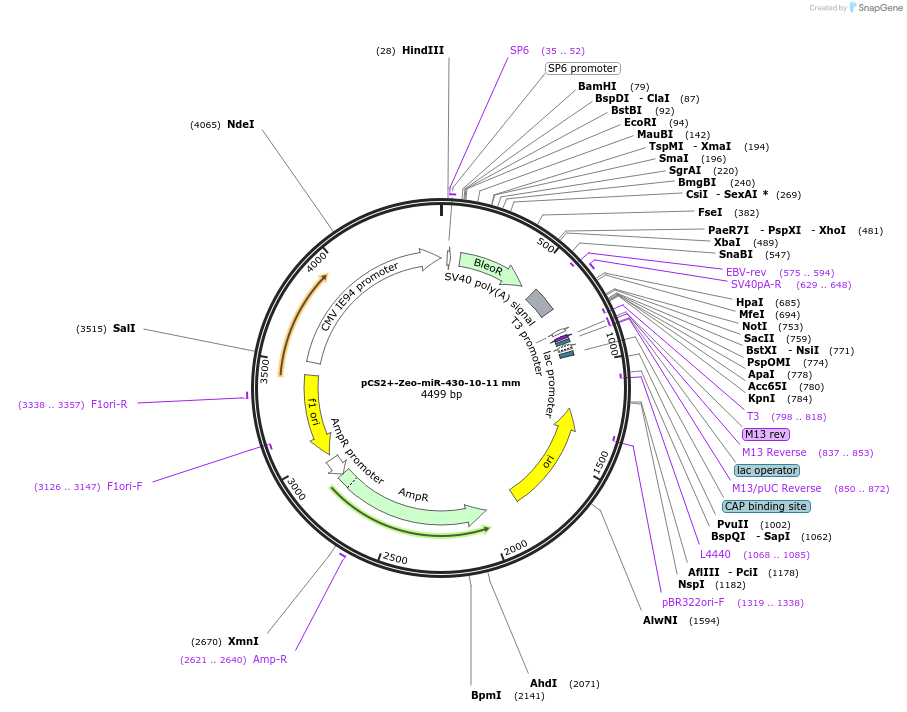
pCS2+-Zeo-miR-430-10-11 mm
Plasmid#221727PurposeExpression of Zeocin resistance marker followed by miR-430 target site insertion with mismatches at position 10 and 11 in zebrafish embryos.DepositorInsertBleoR-miR-430-10-11 mm
UseFish expressionMutationMismatch at positions 10 and 11 of miR-430 sitePromotersimian CMV IE94Available SinceDec. 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
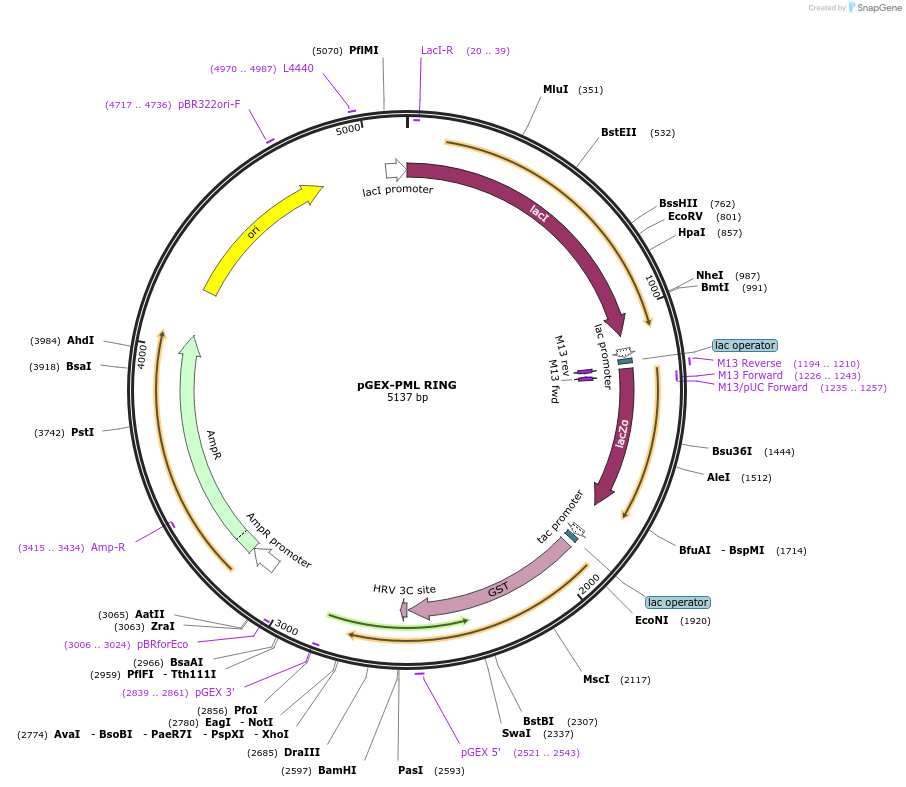
pGEX-PML RING
Plasmid#185631Purposeexpression of GST-tagged PML RING domain in bacterial cellsDepositorAvailable SinceSept. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
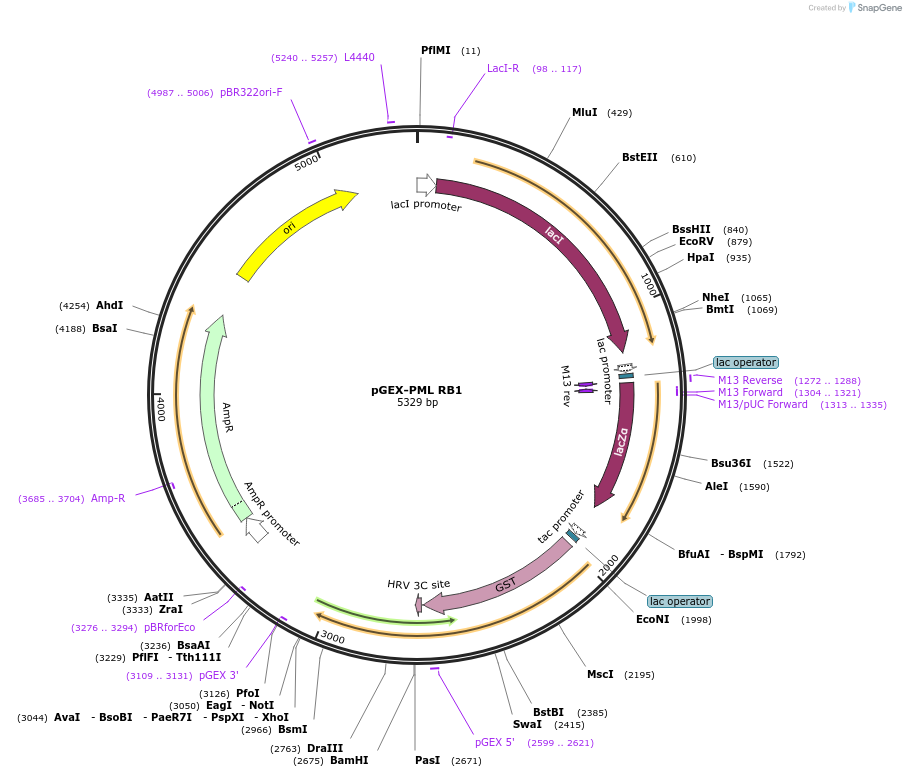
pGEX-PML RB1
Plasmid#185636Purposeexpression of GST-tagged PML fragment 49-168 (RING + B-box1) in bacterial cellsDepositorAvailable SinceSept. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
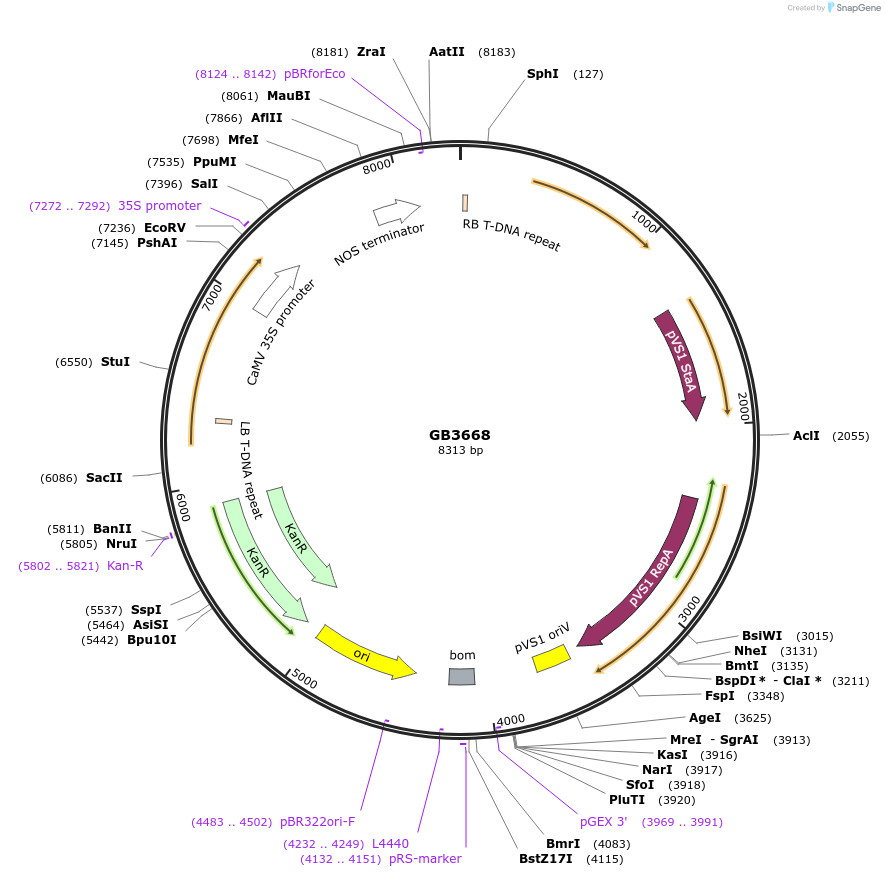
GB3668
Plasmid#215243PurposeTU for the expression of mAID (N-tag): AcrIIA4 w/o NLS protein Codon optimized for N. benthamiana expression under the regulation of the 35S promoter and TNOS terminatorDepositorInsertP35S:mAID:AcrIIA4:TNOS
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJune 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
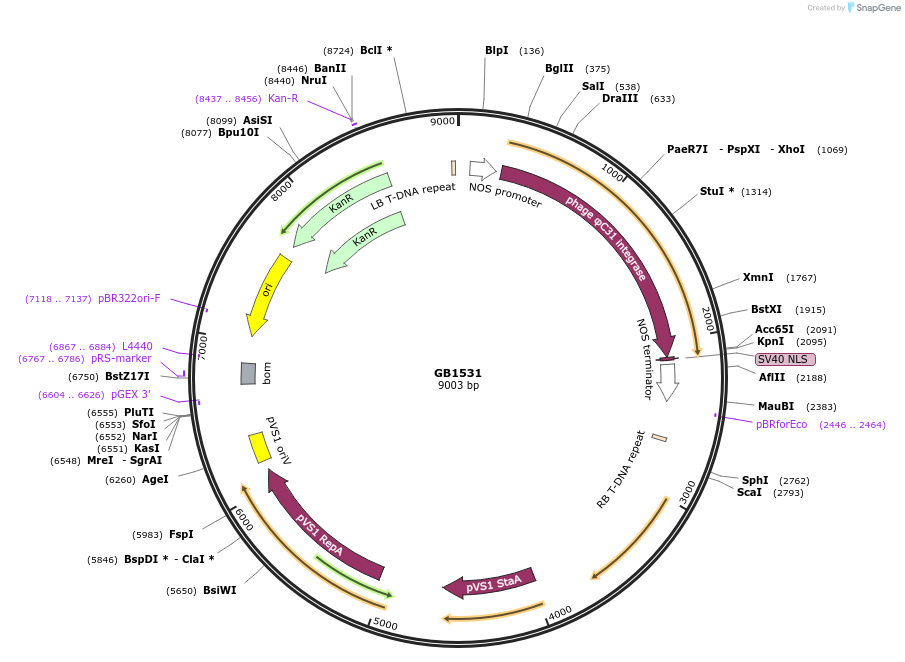
GB1531
Plasmid#215242PurposeTU for constitutive expression of Streptomyces phage phiC31 integrase. Catalyzes site-specific recombination between phiC31 attB and attP sites. Contains SV40 NLS. N. benthamiana codon optimized.DepositorInsertPNos:PhiC31:TNos
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJune 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
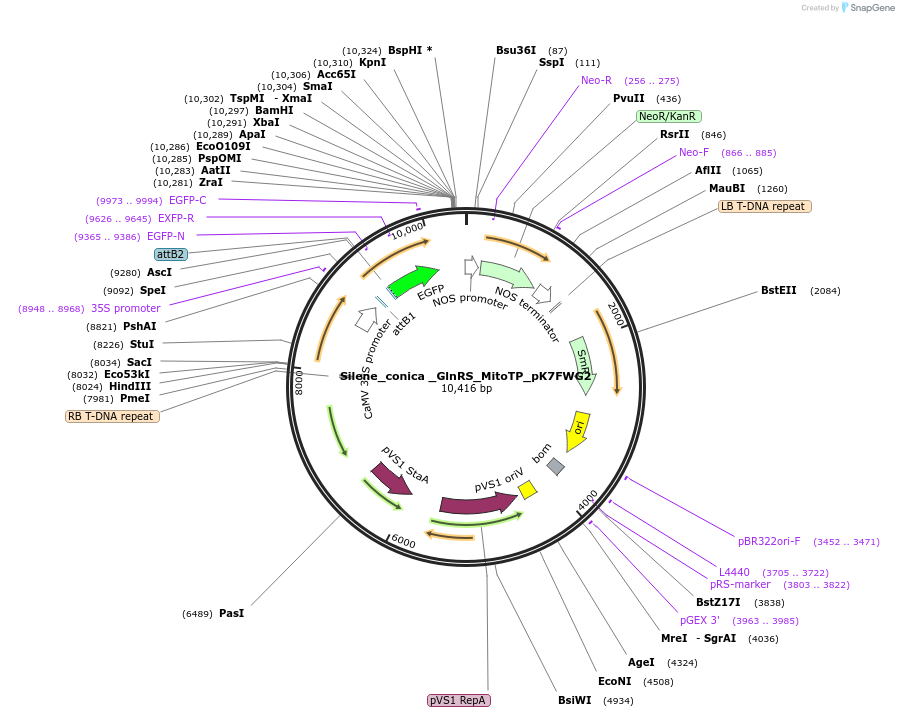
Silene_conica _GlnRS_MitoTP_pK7FWG2
Plasmid#202654PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from Silene conica cytosolic GlnRS predicted to be re-targeted to mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
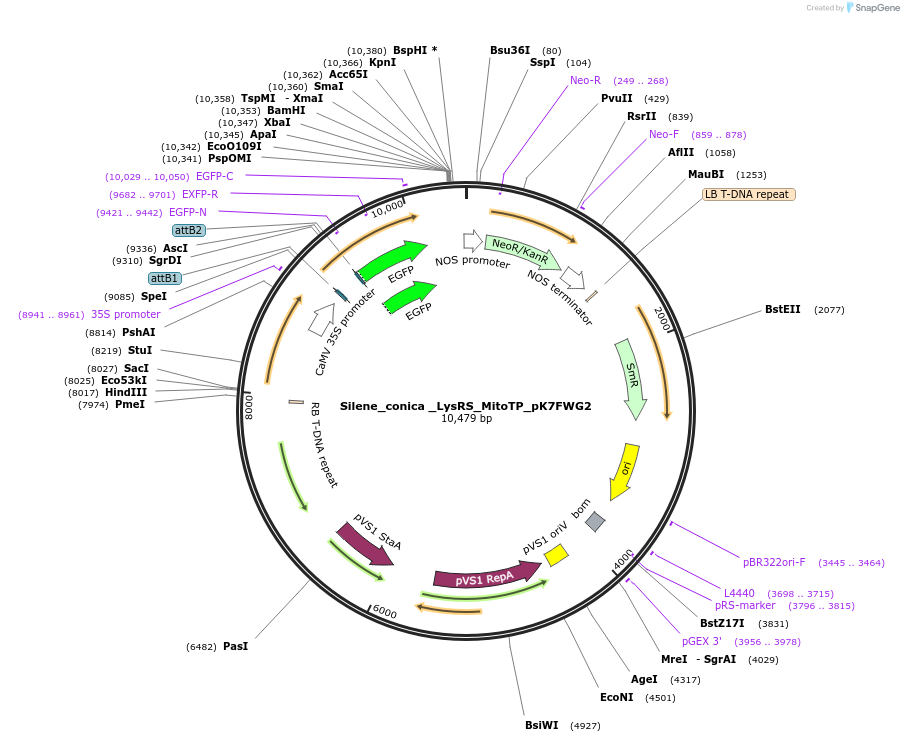
Silene_conica _LysRS_MitoTP_pK7FWG2
Plasmid#202655PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from Silene conica cytosolic LysRS predicted to be re-targeted to mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
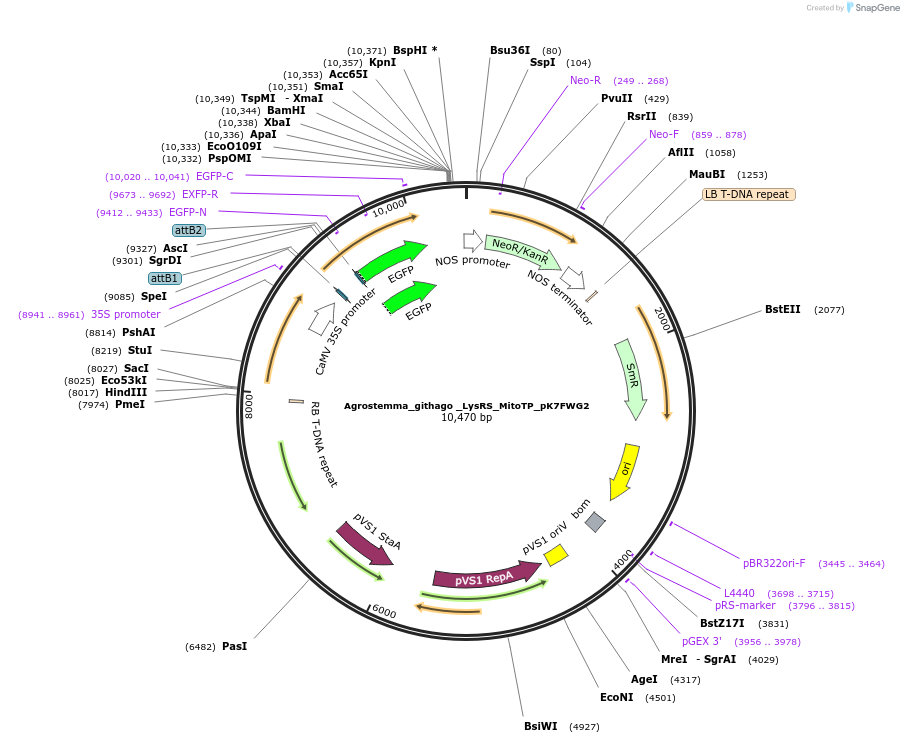
Agrostemma_githago _LysRS_MitoTP_pK7FWG2
Plasmid#202656PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from Agrostemma githago cytosolic LysRS predicted to be re-targeted to mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
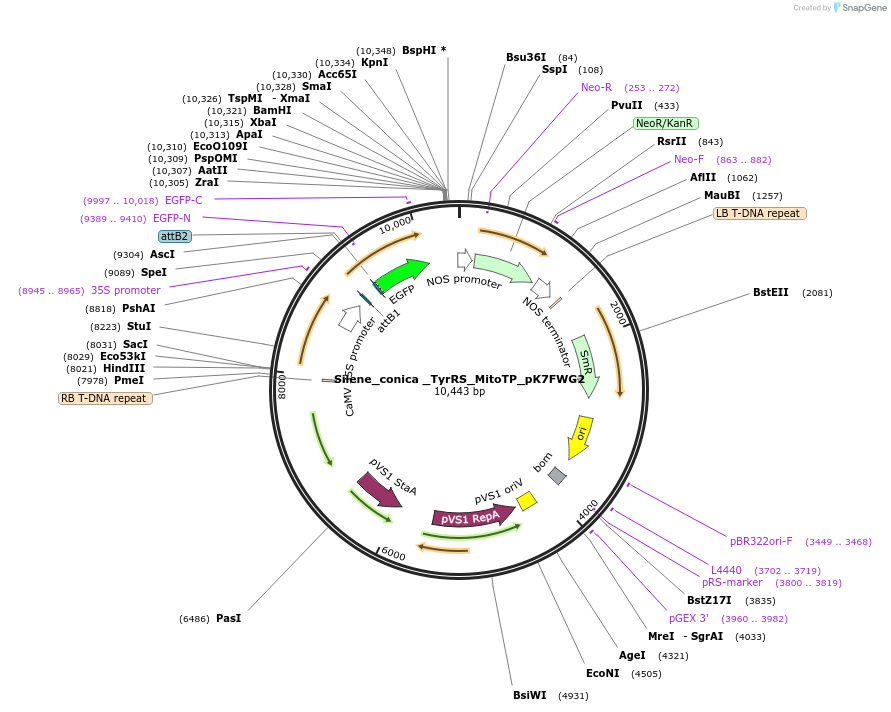
Silene_conica _TyrRS_MitoTP_pK7FWG2
Plasmid#202657PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from Silene conica cytosolic TyrRS predicted to be re-targeted to mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
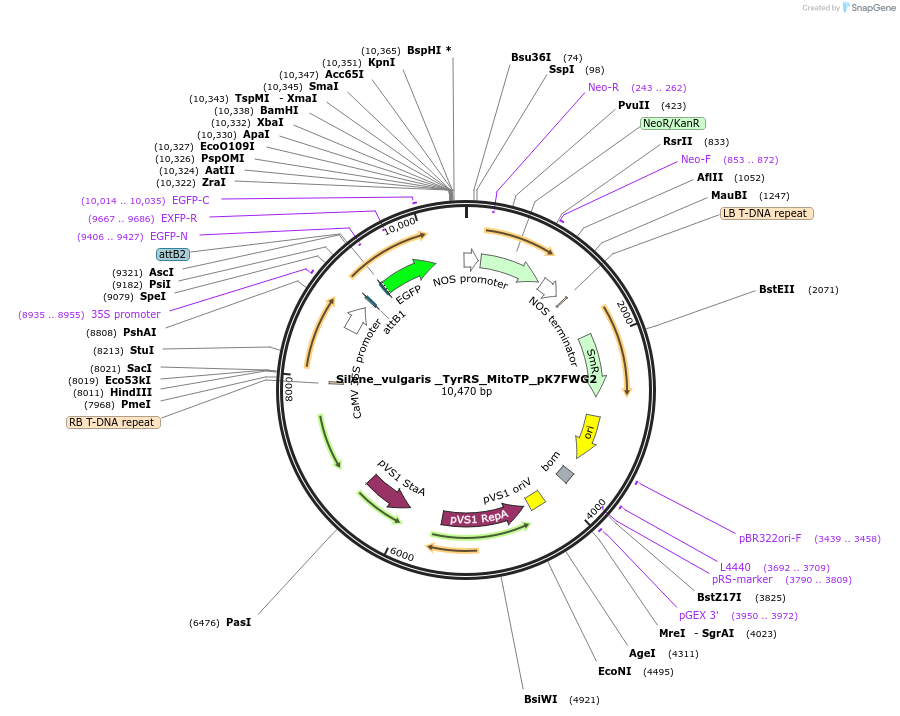
Silene_vulgaris _TyrRS_MitoTP_pK7FWG2
Plasmid#202658PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from Silene vulgaris cytosolic TyrRS predicted to be re-targeted to mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits