We narrowed to 5,240 results for: codon optimized
-
Plasmid#232055PurposeExpresses construct for surface tethering: R. norvegicus NFL tail domainDepositorInsertNeurofilament-Light tail domain
Tags6xHisExpressionBacterialMutationcDNA codon-optimized for E. coli; added N-termina…PromoterT7Available SinceMarch 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
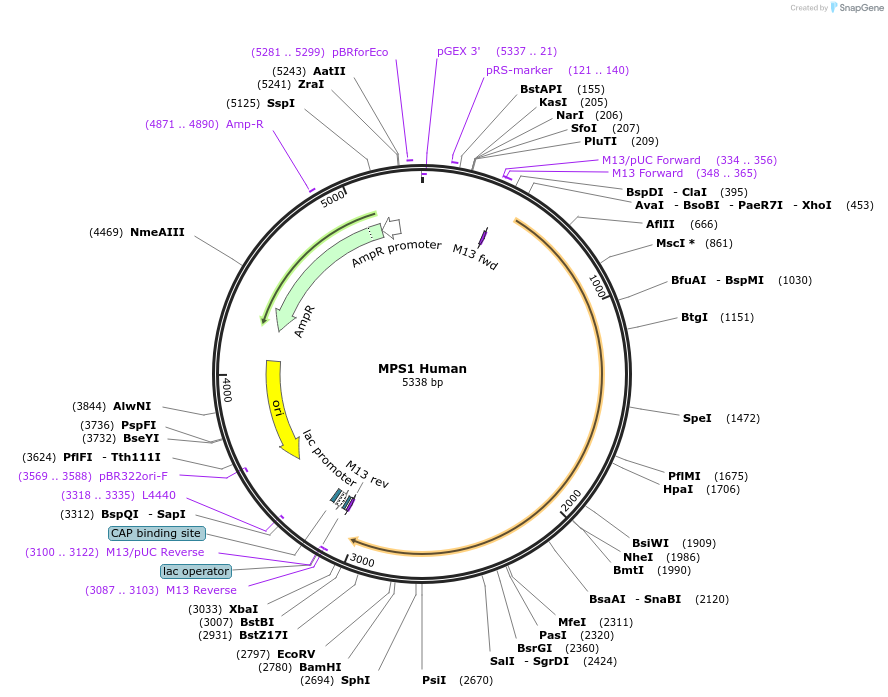
MPS1 Human
Plasmid#232149PurposeMPS1 from Human codon optimized for S. cerevisiae in pUCIDTDepositorInsertMPS1 (TTK Human)
ExpressionYeastAvailable SinceFeb. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
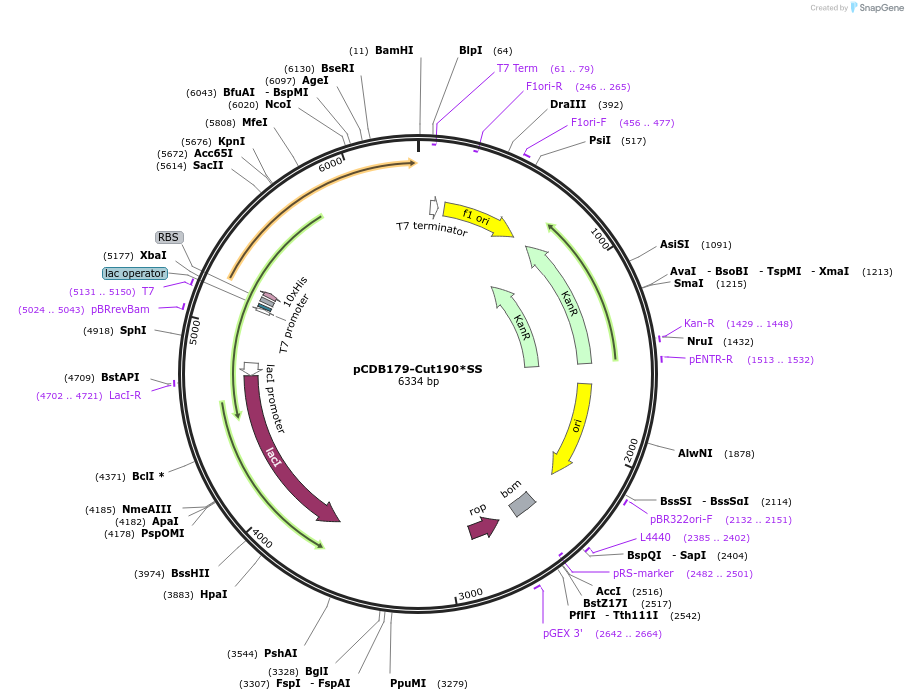
pCDB179-Cut190*SS
Plasmid#218706PurposeEncodes codon optimized Cut190*SSenzyme for bacterial expressionDepositorInsertCut190*SS
Tags10xHisExpressionBacterialAvailable SinceNov. 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
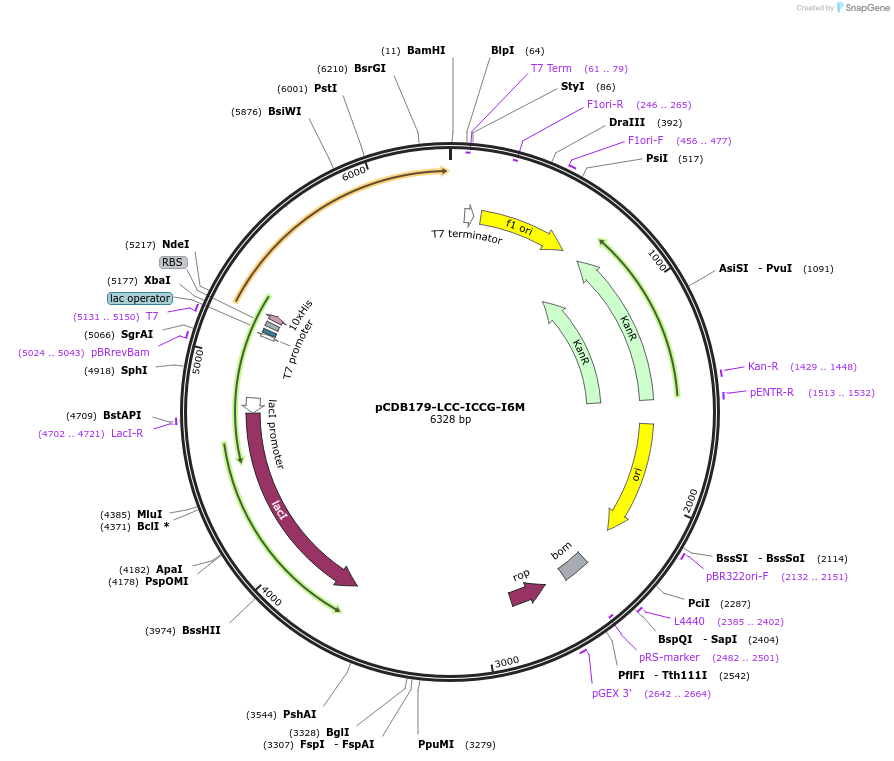
pCDB179-LCC-ICCG-I6M
Plasmid#218690PurposeEncodes codon optimized LCC-ICCG I6M enzyme for bacterial expressionDepositorInsertLCC-ICCG
Tags10xHisExpressionBacterialAvailable SinceNov. 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
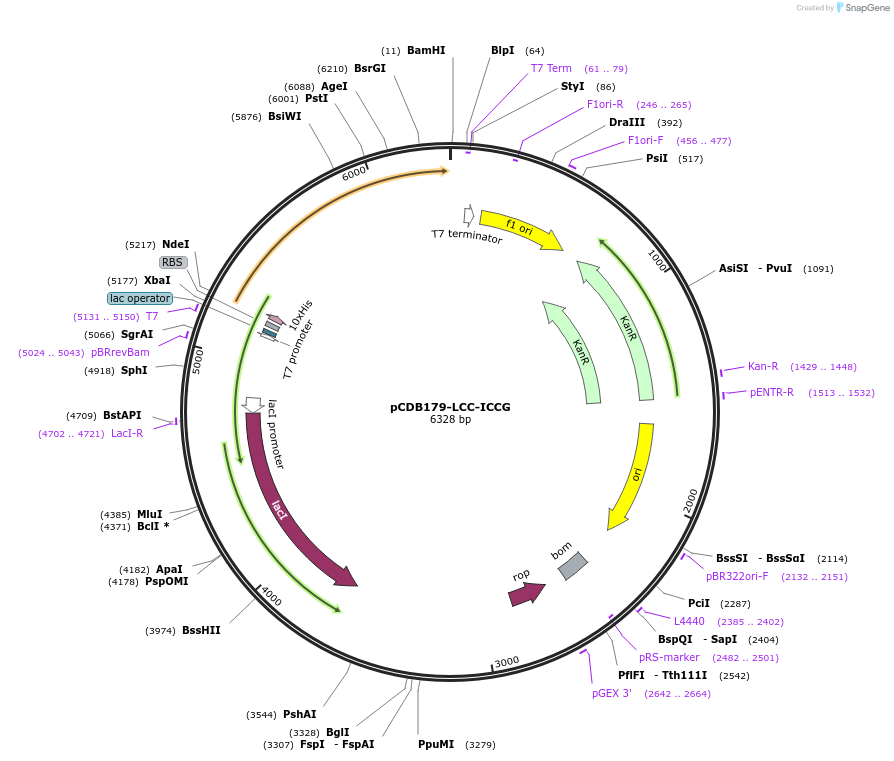
pCDB179-LCC-ICCG
Plasmid#218687PurposeEncodes codon optimized LCC-ICCG enzyme for bacterial expressionDepositorInsertLCC-ICCG
Tags10xHisExpressionBacterialAvailable SinceNov. 7, 2024AvailabilityAcademic Institutions and Nonprofits only -
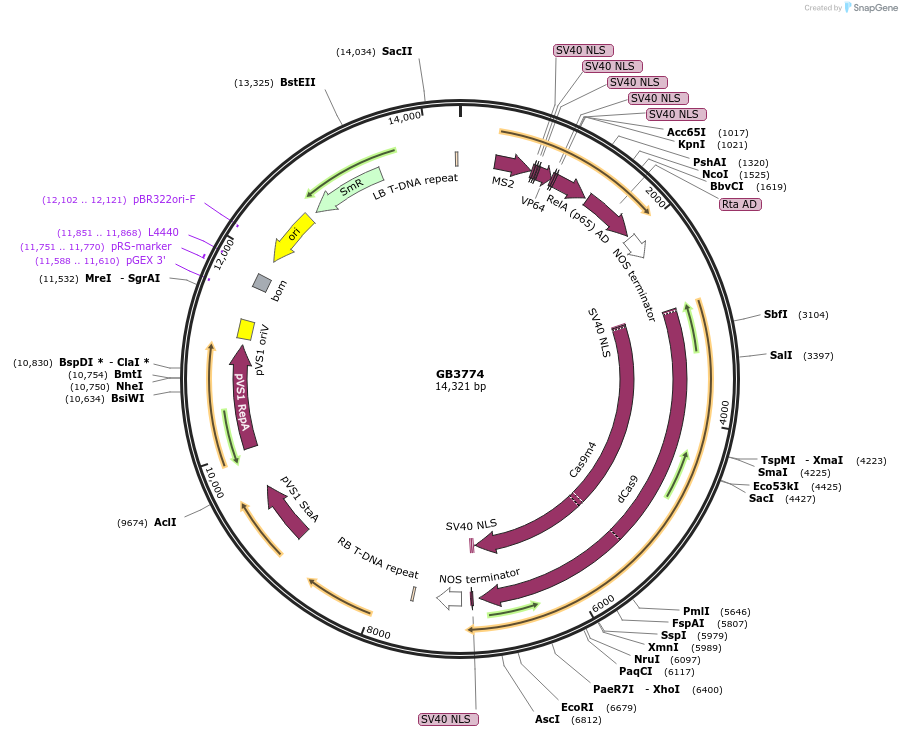
GB3774
Plasmid#215247PurposeCDS of CPH from N. nambi codon optimized for N.benthamiana. This gene is the final step and recycling of the bioluminiscente pathwayDepositorInsertCPH
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceOct. 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
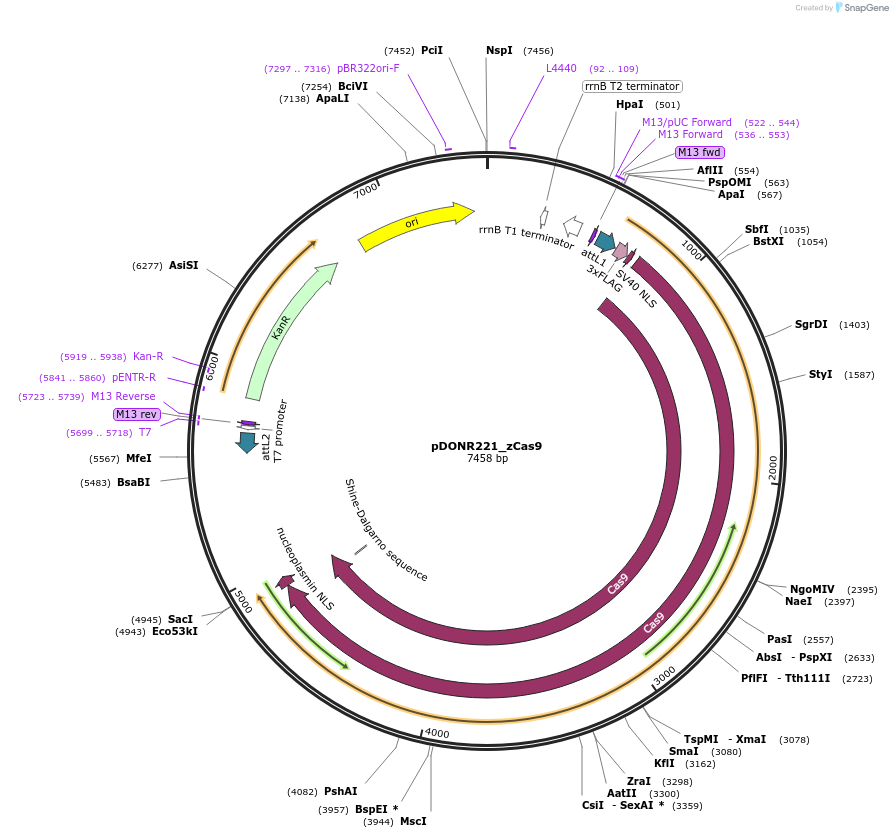
pDONR221_zCas9
Plasmid#213913PurposeEntry clone containing 3×FLAG-NLS-zCas9-NLS encoding sequence, Pisum sativum rbcS E9 terminator. For use in plants and compatible with the MultiSite Gateway systemDepositorInsertzCas9
UseCRISPRMutationZea mays codon-optimized Cas9Available SinceJuly 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
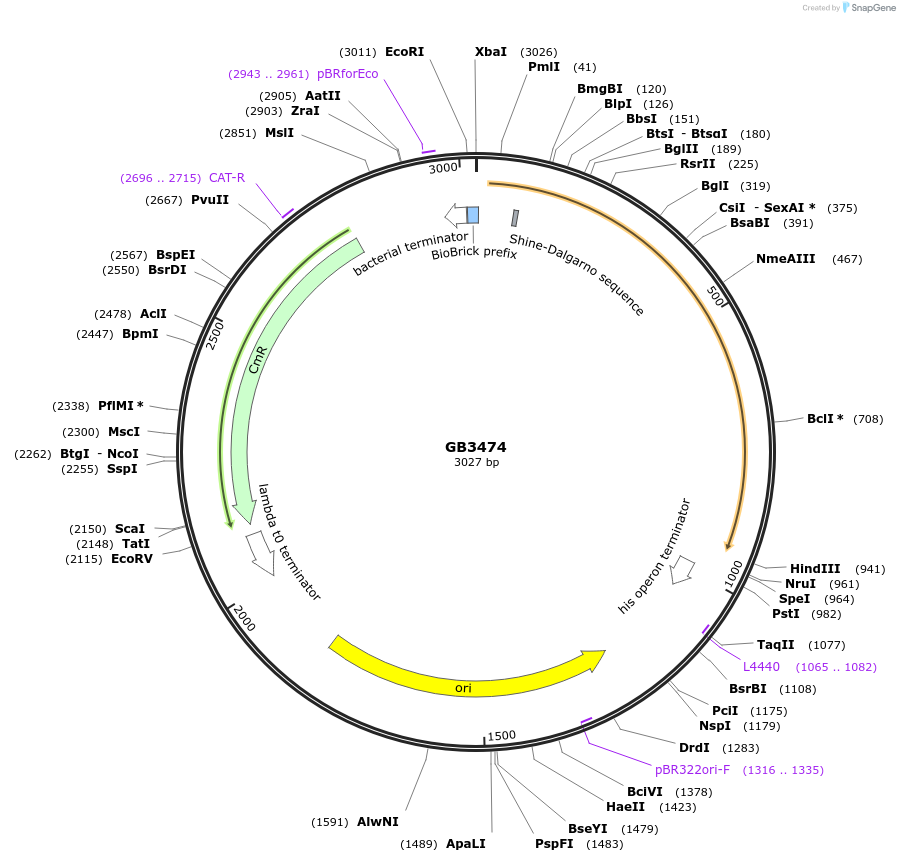
GB3474
Plasmid#215370PurposeCDS of CPH from N. nambi codon optimized for N.benthamiana. This gene is the final step and recycling of the bioluminiscente pathwayDepositorInsertCPH
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJune 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
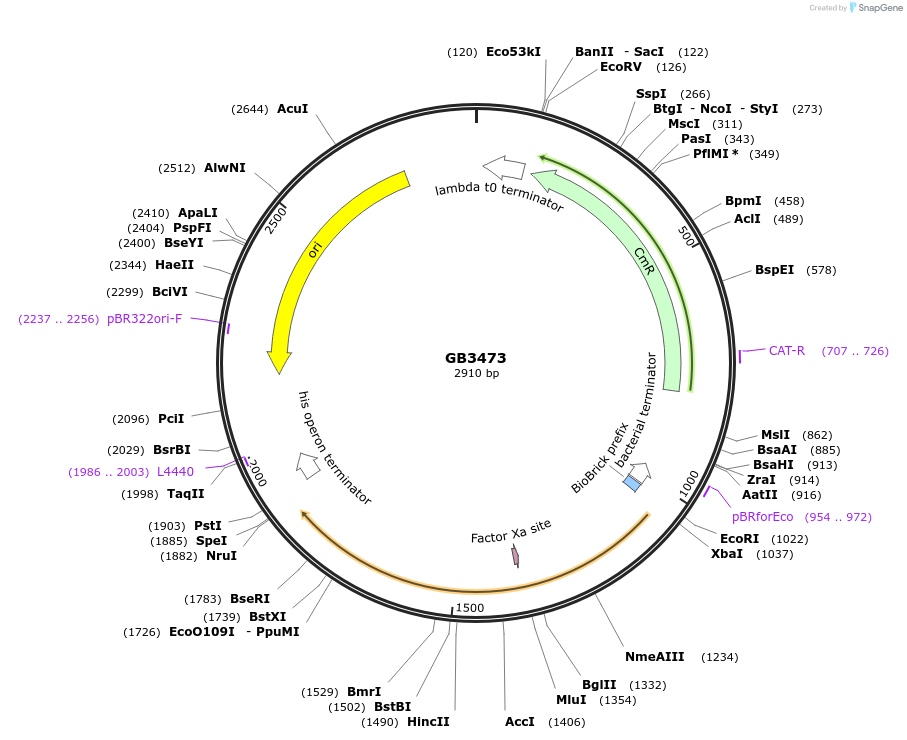
GB3473
Plasmid#215246PurposeCDS of Luz gene from N. nambi codon optimized for N.benthamiana. This gene generate the luminiscence of the bioluminiscente pathwayDepositorInsertLuz
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJune 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
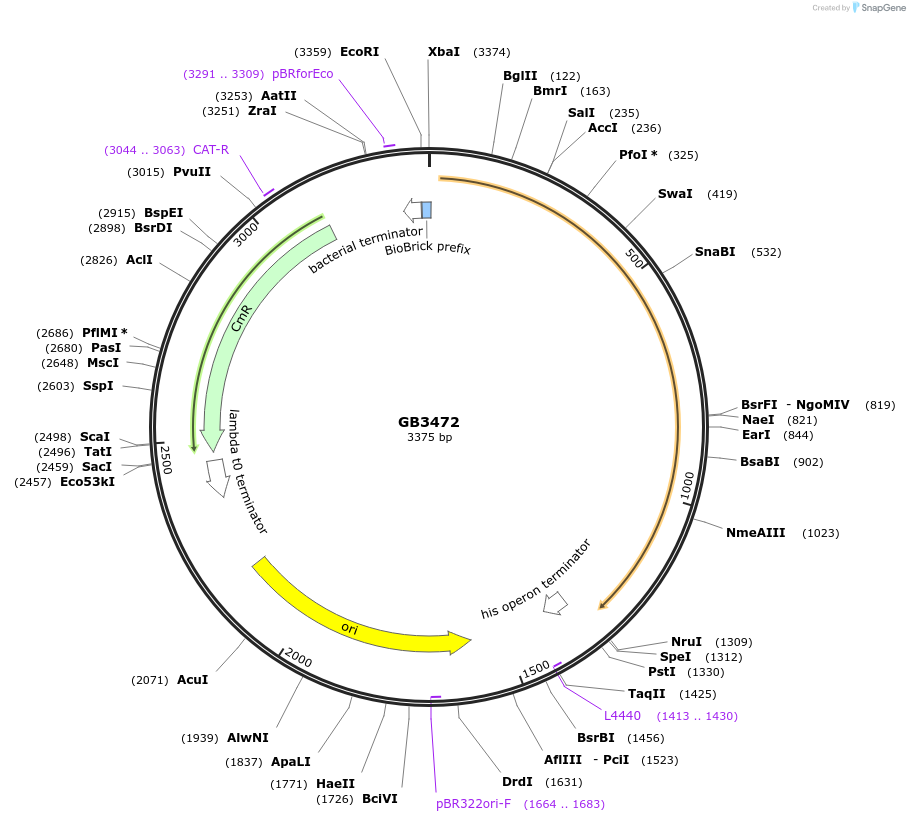
GB3472
Plasmid#215245PurposeCDS of H3H from N. nambi codon optimized for N.benthamiana. This gene is the second step of the bioluminiscente pathwayDepositorInsertH3H
UseSynthetic BiologyMutationBsaI and BsmBI sites removedAvailable SinceJune 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
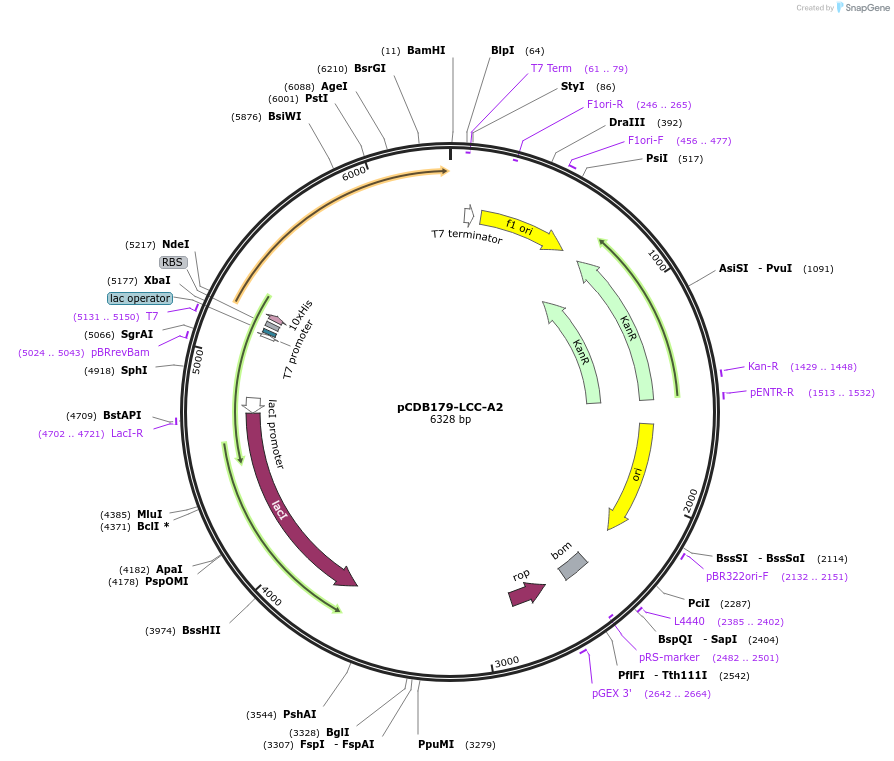
pCDB179-LCC-A2
Plasmid#218691PurposeEncodes codon optimized LCC-A2 enzyme for bacterial expressionDepositorInsertLCC-A2
Tags10xHisExpressionBacterialAvailable SinceMay 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
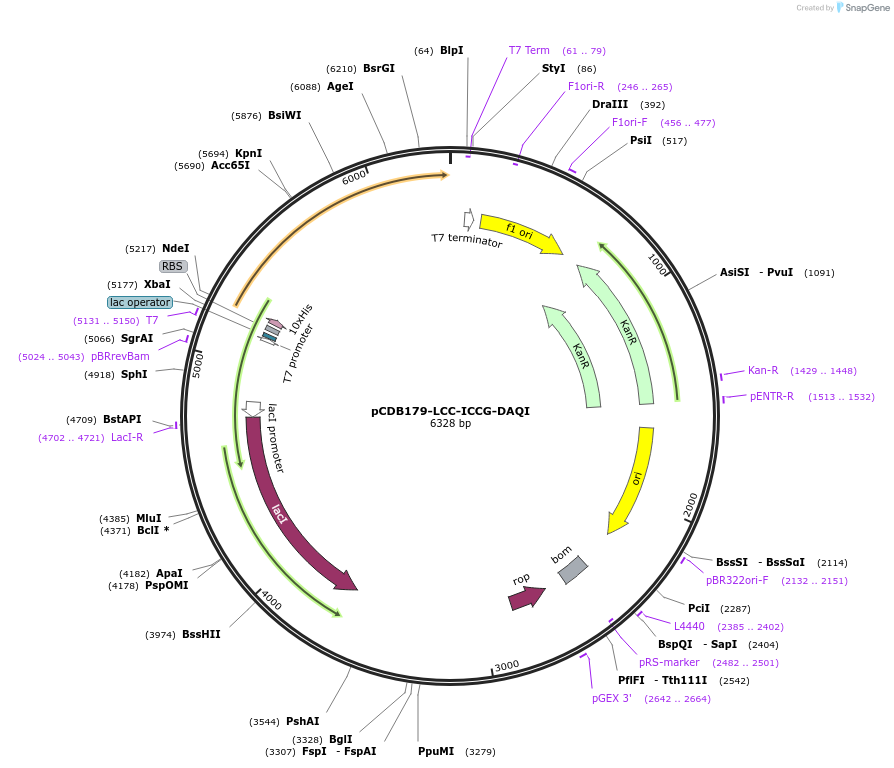
pCDB179-LCC-ICCG-DAQI
Plasmid#218689PurposeEncodes codon optimized LCC-ICCG DAQI enzyme for bacterial expressionDepositorInsertLCC-ICCG-DAQI
Tags10xHisExpressionBacterialAvailable SinceMay 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
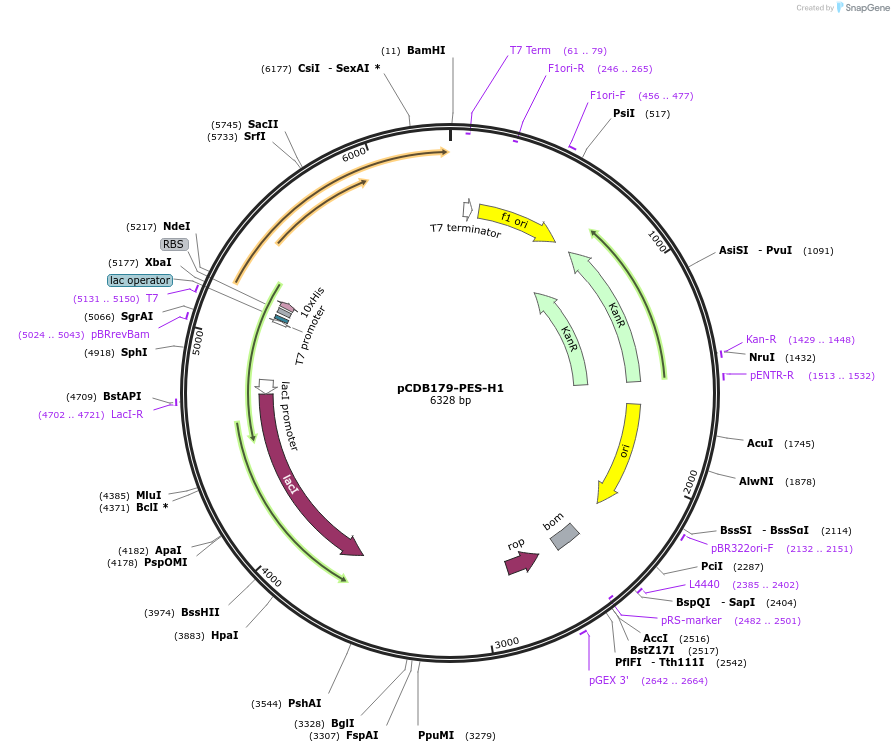
pCDB179-PES-H1
Plasmid#218704PurposeEncodes codon optimized PES-H1 enzyme for bacterial expressionDepositorInsertPES-H1
Tags10xHisExpressionBacterialAvailable SinceMay 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
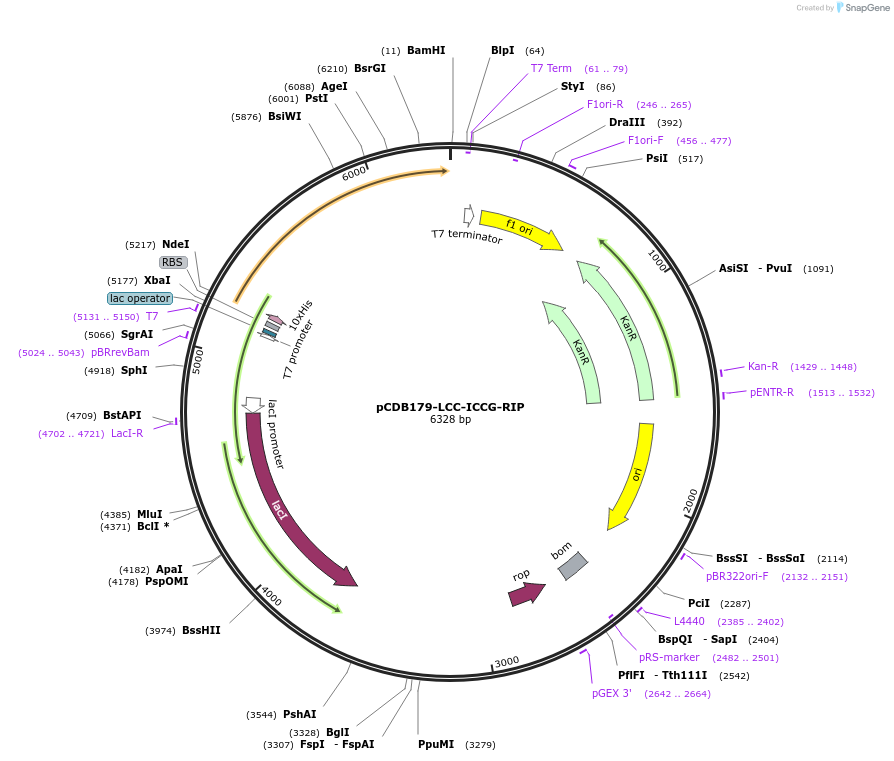
pCDB179-LCC-ICCG-RIP
Plasmid#218688PurposeEncodes codon optimized LCC-ICCG RIP enzyme for bacterial expressionDepositorInsertLCC-ICCG-RIP
Tags10xHisExpressionBacterialAvailable SinceMay 21, 2024AvailabilityAcademic Institutions and Nonprofits only -
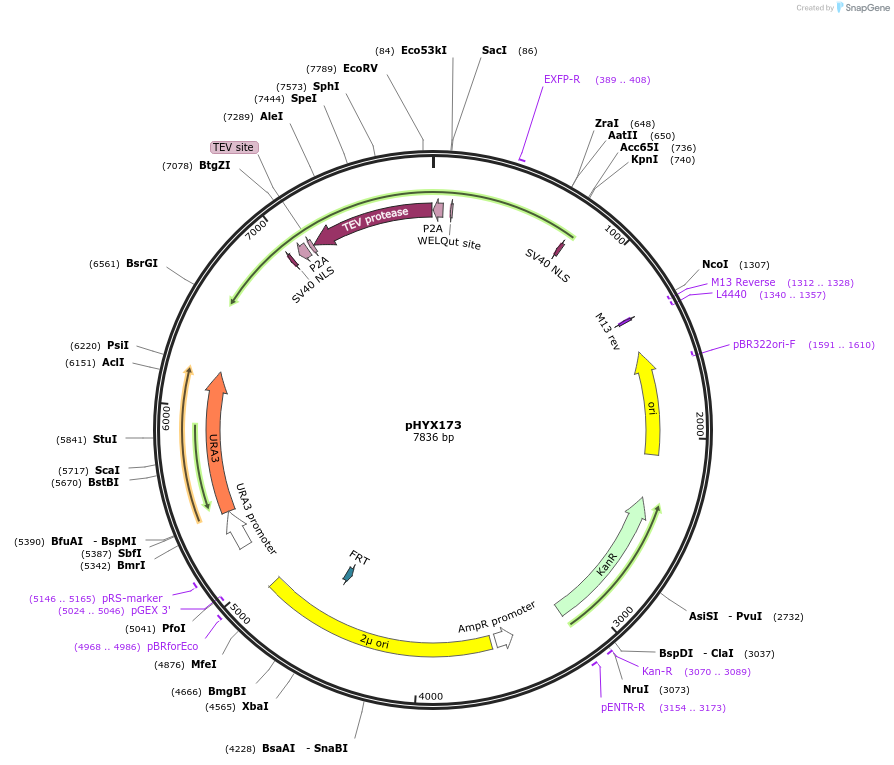
pHYX173
Plasmid#202817PurposePlasmid expressing the TEV protease, codon-optimized for Saccharomyces cerevisiae.DepositorInsertTEV protease
ExpressionYeastPromoterScPCK1Available SinceNov. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
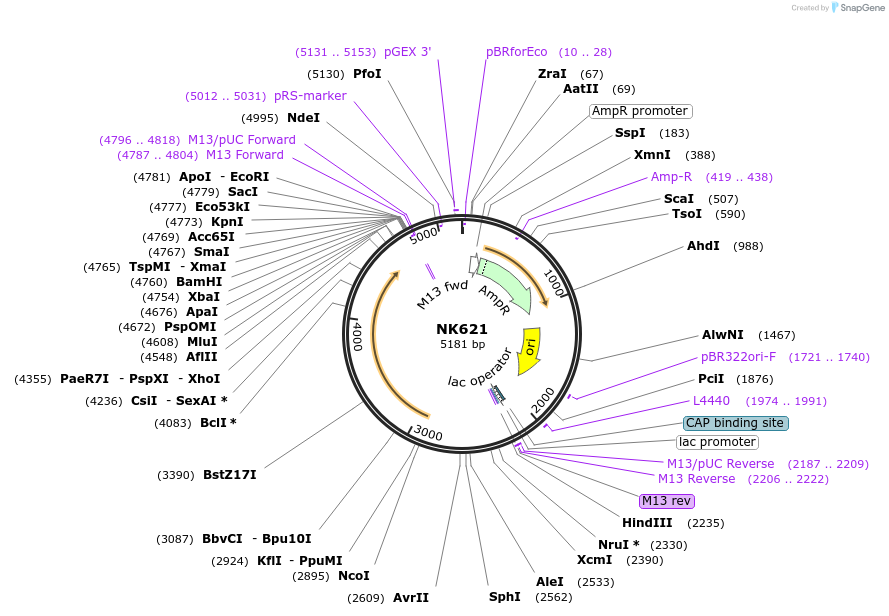
NK621
Plasmid#196405PurposeStrong expression of firefly luciferase codon-optimized for S. rosettaDepositorInsertSro5actin-firefly-Sro3actin
UseLuciferaseMutationN/A (synthetic)Available SinceAug. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
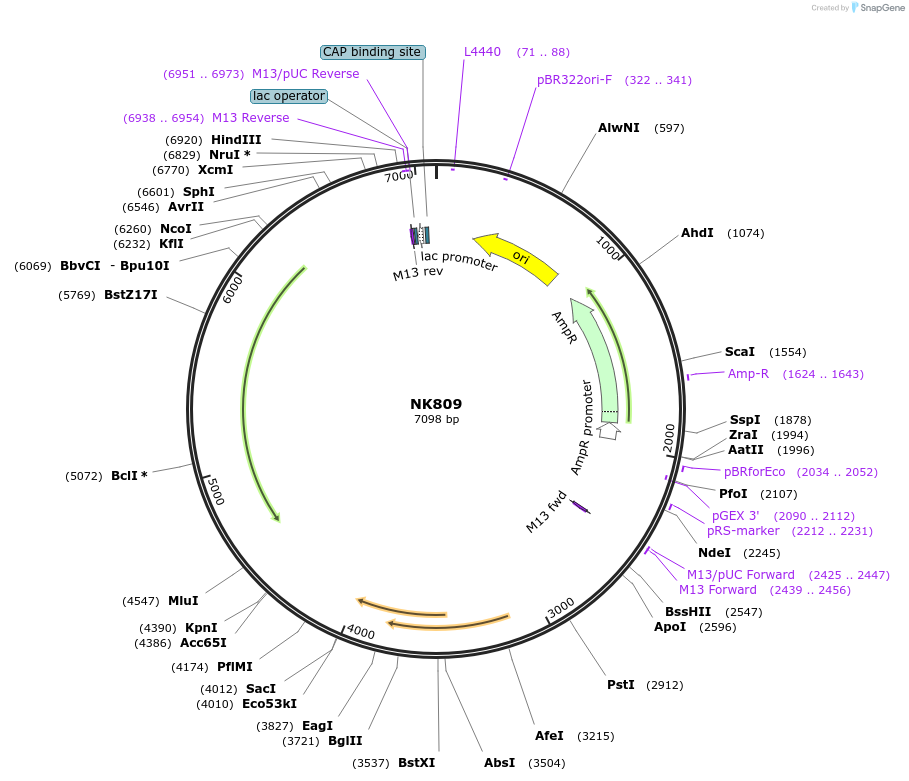
NK809
Plasmid#196406PurposeStrong expression of firefly and nanoluc luciferases codon-optimized for S. rosettaDepositorInsertSro5EFL-nanoluc-Sro3EFL/Sro5actin-firefly-Sro3actin
UseLuciferaseMutationN/A (synthetic)Available SinceAug. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
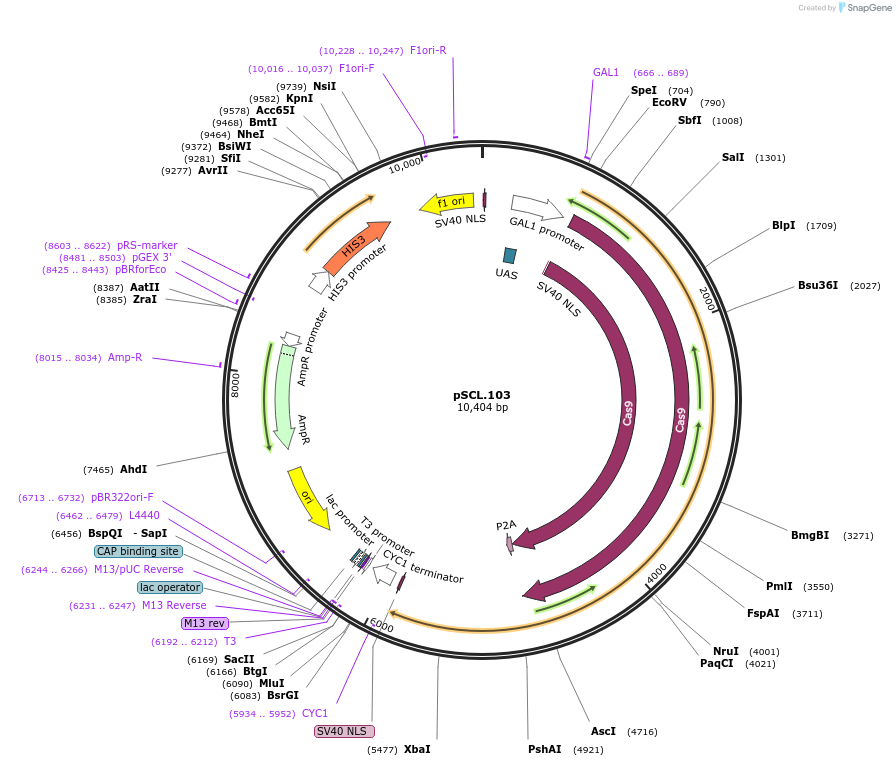
pSCL.103
Plasmid#184988PurposeExpress Cas9-P2A-Eco1RTDepositorInsertCas9-P2A-Eco1RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
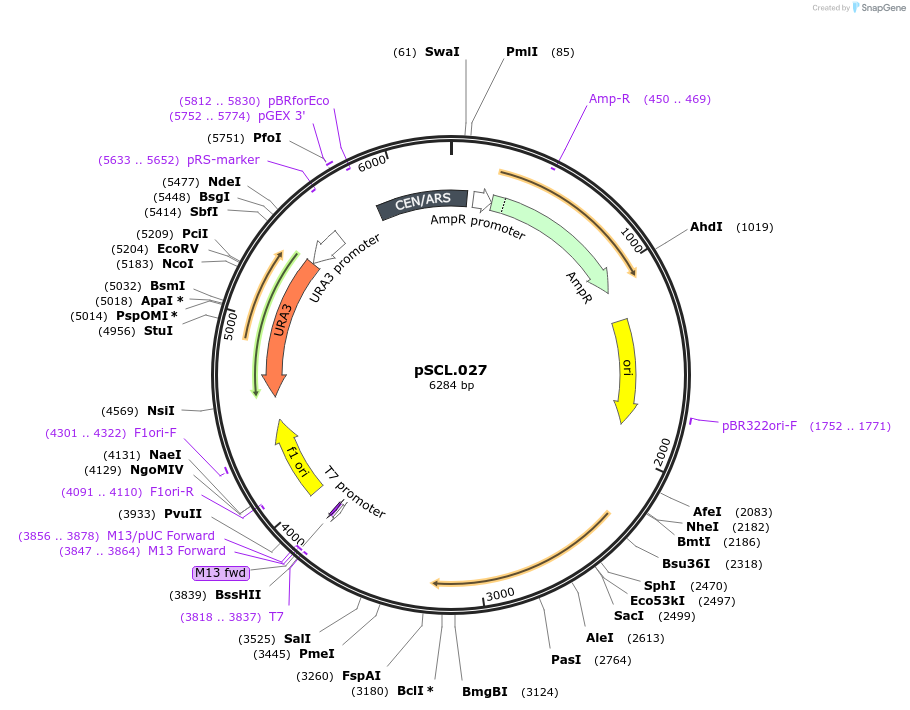
pSCL.027
Plasmid#184964PurposeExpress -Eco1 RT and ncRNA in yeastDepositorInsertEco1: RT and ncRNA(wt), a1/a2 length: 12
ExpressionYeastMutationhuman codon optimized RTPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
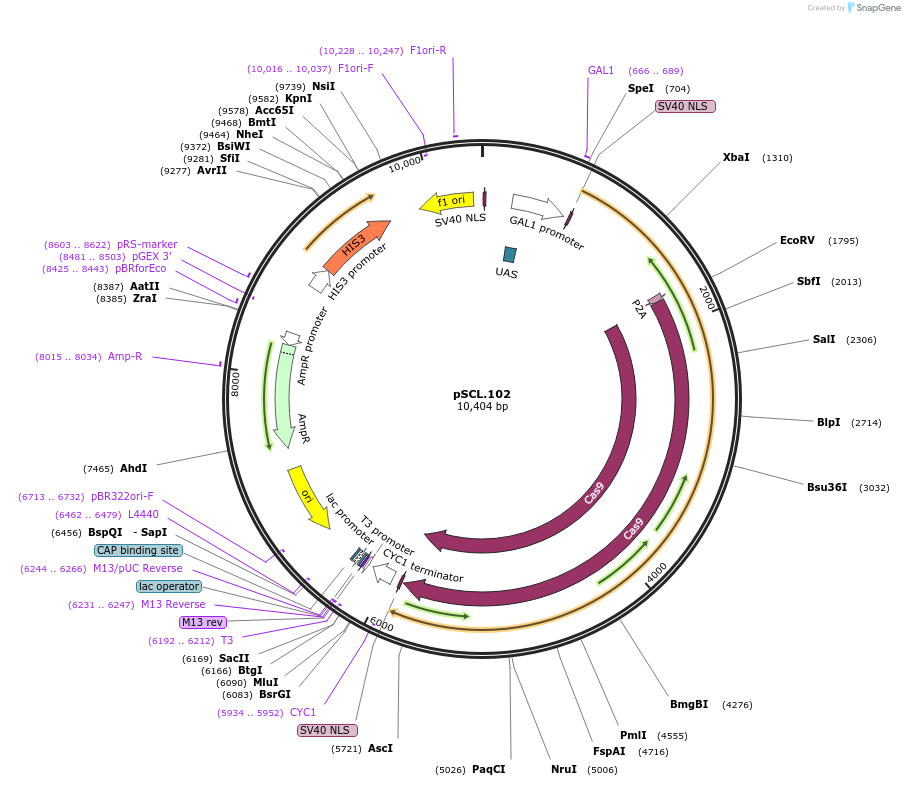
pSCL.102
Plasmid#184987PurposeExpress -Eco1 RT-P2A-Cas9DepositorInsertEco1RT-P2A-Cas9
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only