We narrowed to 11,776 results for: Vars
-
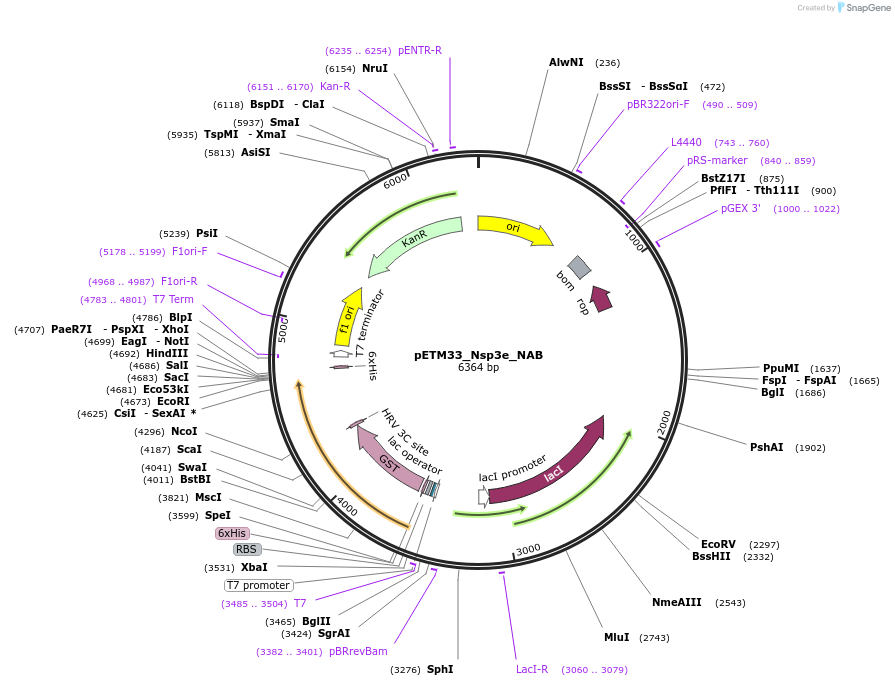
Plasmid#156473PurposeBacterial expression of Sars-CoV2 Nsp3e_NAB protein with His-tag and GST-tagDepositorInsertNsp3e_NAB (ORF1ab Severe acute respiratory syndrome coronavirus 2, Synthetic)
TagsHis-GSTExpressionBacterialMutationGene insert is codon optimized for Ecoli.PromoterT7/LacOAvailable SinceAug. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
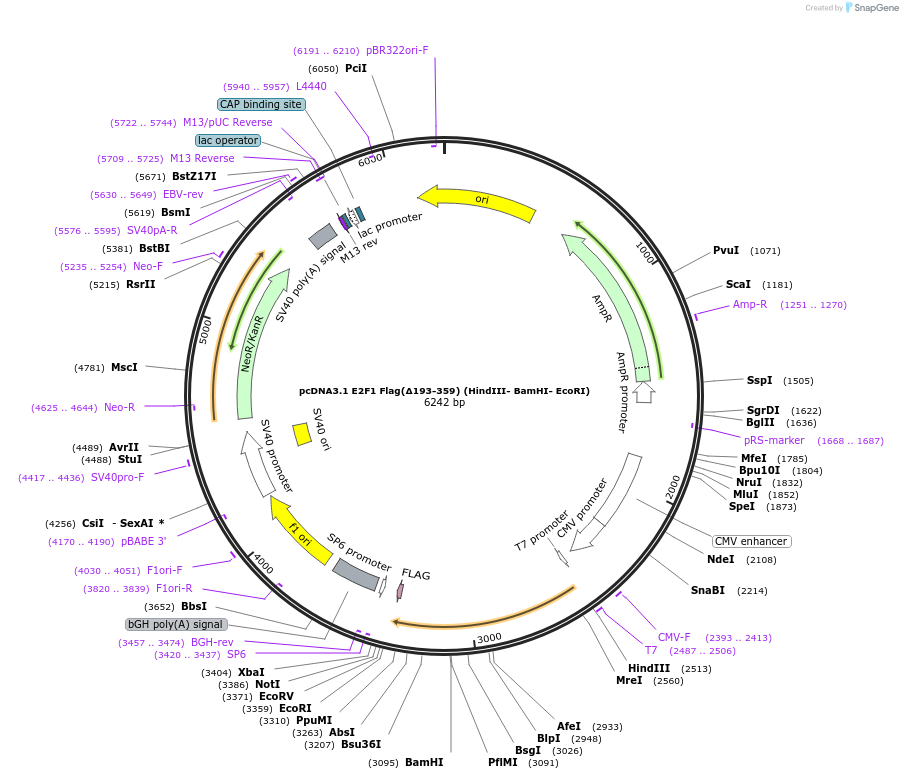
pcDNA3.1 E2F1 Flag(Δ193-359) (HindIII- BamHI- EcoRI)
Plasmid#70667PurposeHuman mutant of E2F1 lacking amino acids 193-359DepositorInsertE2F1 lacking residues 193 to 359 (E2F1 Human)
TagsFLAGExpressionMammalianMutationlacks amino acids 193-359PromotercmvAvailable SinceMay 16, 2017AvailabilityAcademic Institutions and Nonprofits only -
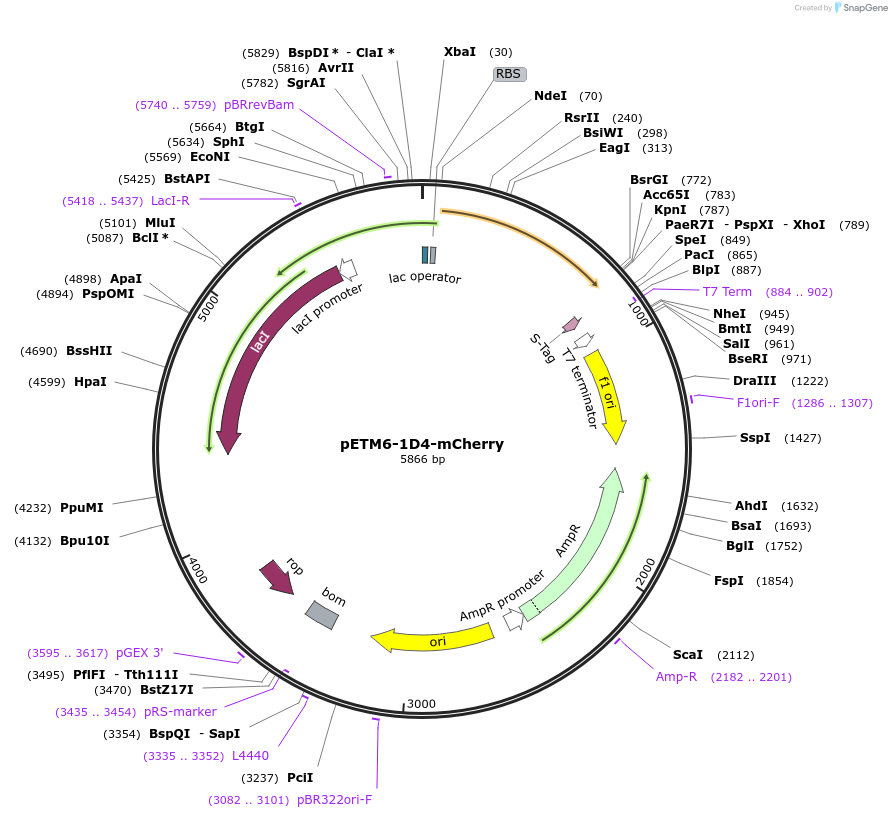
pETM6-1D4-mCherry
Plasmid#73423PurposeReporter plasmid encoding mCherry, codon optimized for E. coli, transcriptionally driven by orthogonal T7-lac promoter variant 1D4.DepositorInsertPromoter 1D4 (orthogonal T7-lac variant)
UseCRISPR and Synthetic BiologyExpressionBacterialPromoterP1D4 (orthogonal T7-lac variant)Available SinceMarch 4, 2016AvailabilityAcademic Institutions and Nonprofits only -
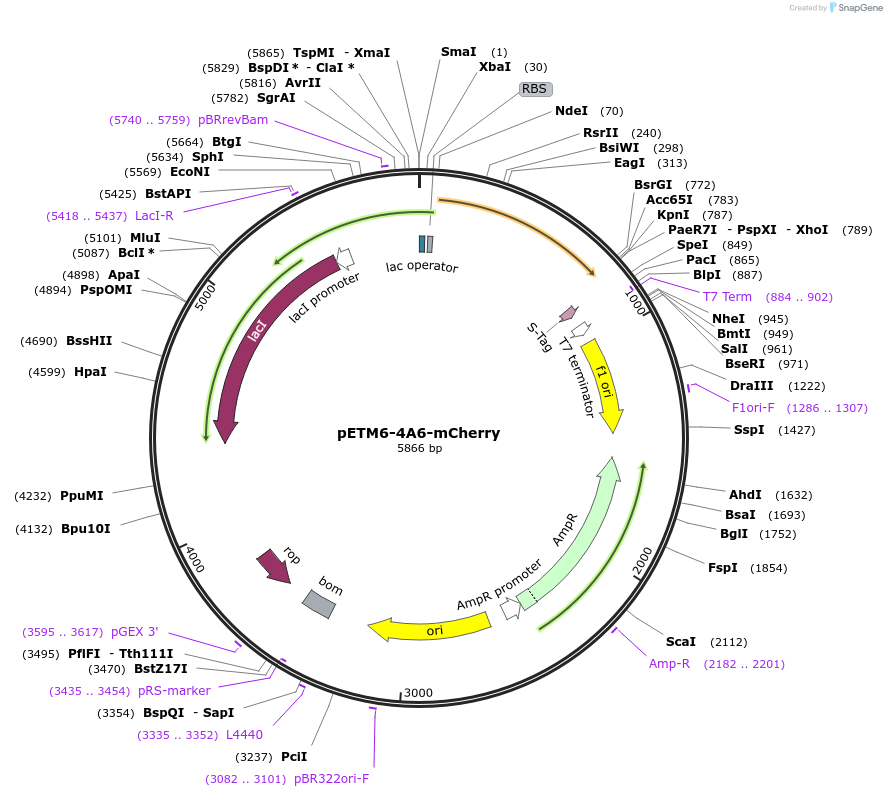
pETM6-4A6-mCherry
Plasmid#73424PurposeReporter plasmid encoding mCherry, codon optimized for E. coli, transcriptionally driven by orthogonal T7-lac promoter variant 4A6.DepositorInsertPromoter 4A6 (orthogonal T7-lac variant)
UseCRISPR and Synthetic BiologyExpressionBacterialPromoterP4A6 (orthogonal T7-lac variant)Available SinceMarch 4, 2016AvailabilityAcademic Institutions and Nonprofits only -
pETM6-1E4-mCherry
Plasmid#73426PurposeReporter plasmid encoding mCherry, codon optimized for E. coli, transcriptionally driven by orthogonal T7-lac promoter variant 1E4.DepositorInsertPromoter 1E4 (orthogonal T7-lac variant)
UseCRISPR and Synthetic BiologyExpressionBacterialPromoterP1E4 (orthogonal T7-lac variant)Available SinceMarch 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
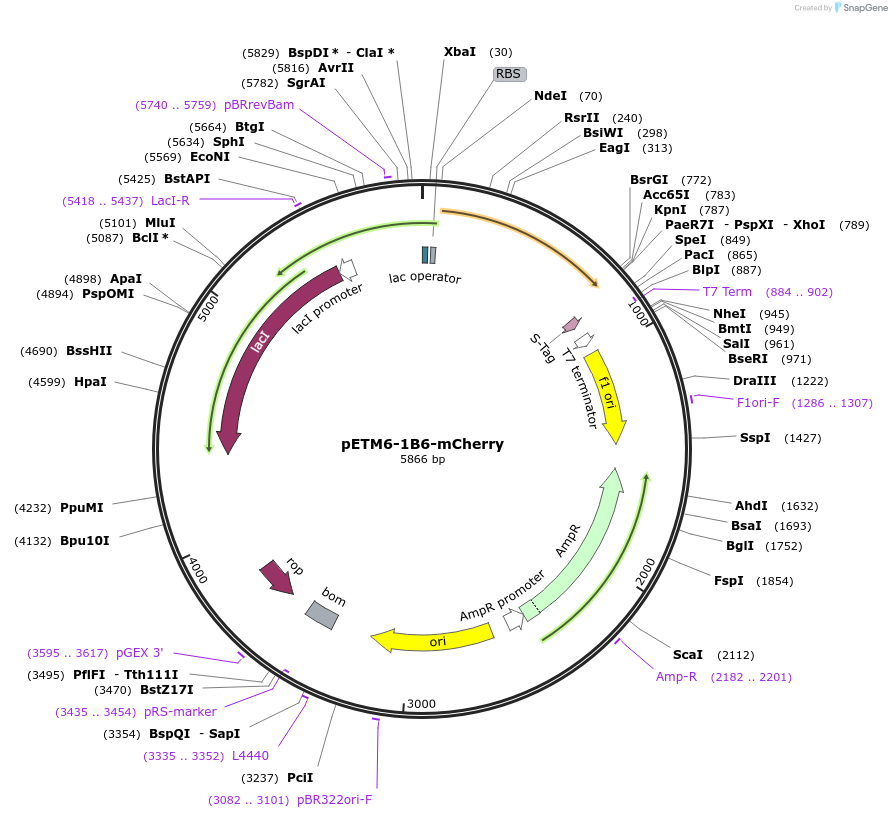
pETM6-1B6-mCherry
Plasmid#73420PurposeReporter plasmid encoding mCherry, codon optimized for E. coli, transcriptionally driven by orthogonal T7-lac promoter variant 1B6.DepositorInsertPromoter 1B6 (orthogonal T7-lac variant)
UseCRISPR and Synthetic BiologyExpressionBacterialPromoterP1B6 (orthogonal T7-lac variant)Available SinceMarch 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
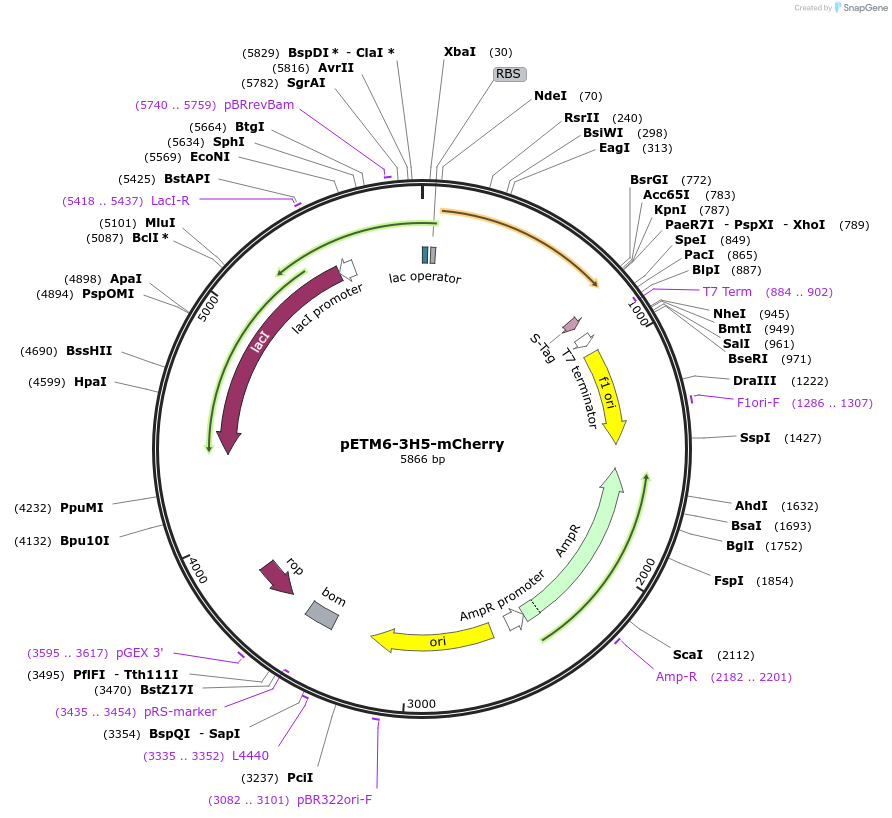
pETM6-3H5-mCherry
Plasmid#73419PurposeReporter plasmid encoding mCherry, codon optimized for E. coli, transcriptionally driven by orthogonal T7-lac promoter variant 3H5.DepositorInsertPromoter 3H5 (orthogonal T7-lac variant)
UseCRISPR and Synthetic BiologyExpressionBacterialPromoterP3H5 (orthogonal T7-lac variant)Available SinceMarch 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
pcDNA3.1 E2F1 Flag(Δ 284−359)(HindIII- BamHI- EcoRI)
Plasmid#70669PurposeHuman mutant of E2F1 lacking amino acids 284 to 359DepositorInsertE2F1 lacking residues 284 to 359 (E2F1 Human)
TagsFlagExpressionMammalianMutationlacks amin acids 284 to 359PromotercmvAvailable SinceNov. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
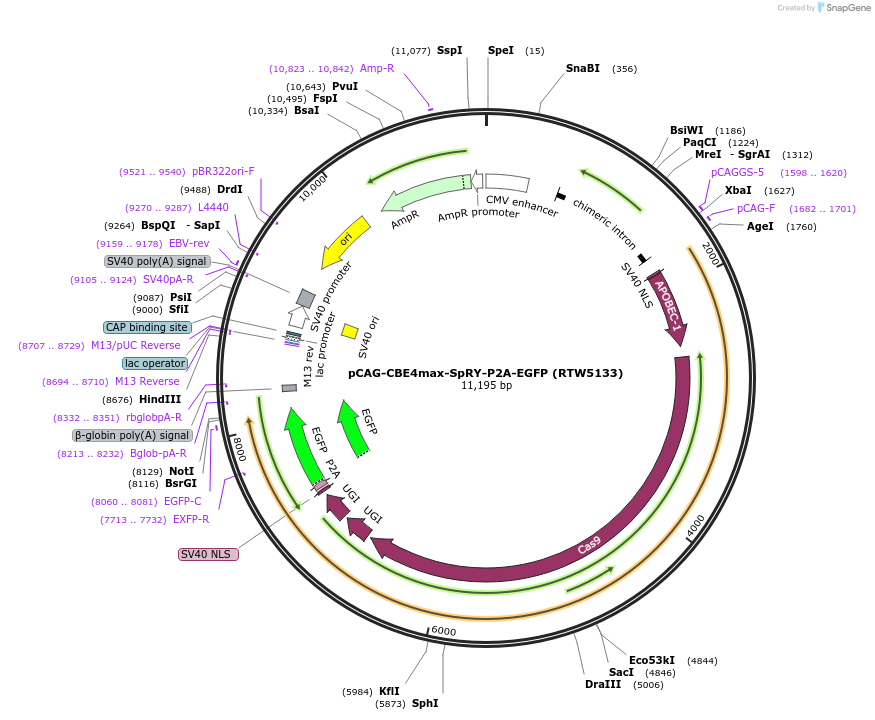
pCAG-CBE4max-SpRY-P2A-EGFP (RTW5133)
Plasmid#139999PurposeCAG promoter expression plasmid for human codon optimized BE4max C-to-T base editor with SpRY(D10A/A61R/L1111R/D1135L/S1136W/G1218K/E1219Q/N1317R/A1322R/R1333P/R1335Q/T1337R) and P2A-EGFPDepositorInserthuman codon optimized CBE4max SpCas9 variant named SpRY with P2A-EGFP
UseCRISPRTagsP2A-EGFPExpressionMammalianMutationnSpCas9=D10A; SpRY=A61R/L1111R/D1135L/S1136W/G121…PromoterCAGAvailable SinceMarch 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
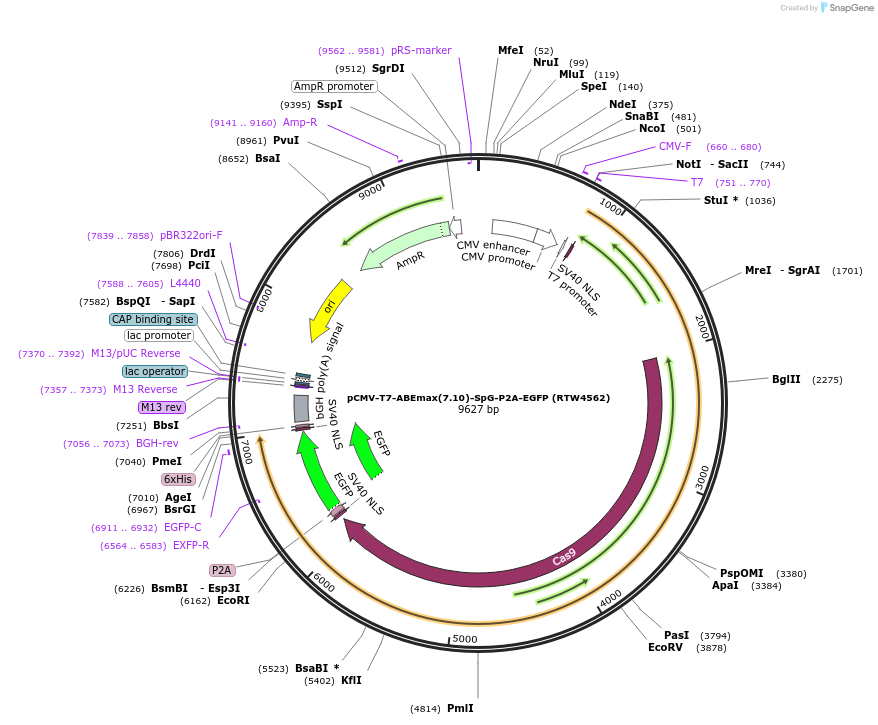
pCMV-T7-ABEmax(7.10)-SpG-P2A-EGFP (RTW4562)
Plasmid#140002PurposeCMV and T7 promoter expression plasmid for human codon optimized ABEmax(7.10) A-to-G base editor with SpG(D10A/D1135L/S1136W/G1218K/E1219Q/R1335Q/T1337R) and P2A-EGFPDepositorInserthuman codon optimized ABEmax(7.10) SpCas9 variant named SpG with P2A-EGFP
UseCRISPR; In vitro transcription; t7 promoterTagsP2A-EGFPExpressionMammalianMutationnSpCas9=D10A; SpG=D1135L/S1136W/G1218K/E1219Q/R13…PromoterCMV and T7Available SinceMarch 26, 2020AvailabilityAcademic Institutions and Nonprofits only